This shows the initial steps of annotation of D. ananassae fosmid 1049B07, including the construction of a gene model for a single gene. This page illustrates the following steps:
| 1. Viewing the fosmid in the GEP UCSC genome browser |
| 2. Finding D. melanogaster orthologs to genes on the fosmid using BLASTX |
| 3. Finding the correct gene symbol for a D. melanogaster ortholog in FlyBase |
| 4. Downloading transcript and CDS information for the D. melanogaster ortholog from the Gene Record Finder |
| 5. Locating the orthologous exons on the fosmid |
| 6. Locating the orthologous CDS on the fosmid |
| 7. Annotating Exon 4, including dealing with a gene on the minus strand |
| 8. Using RNA-Seq data as a tool in annotation |
| 9. Using the Gene Model Checker on a single exon to check our work |
| 10. Comparison of genomic sequences for D. melanogaster and D. ananassae |
| 11. Graphical representation of the D. melanogaster and D. ananassae gene models |
| 12. Species distribution of gain of the intron |
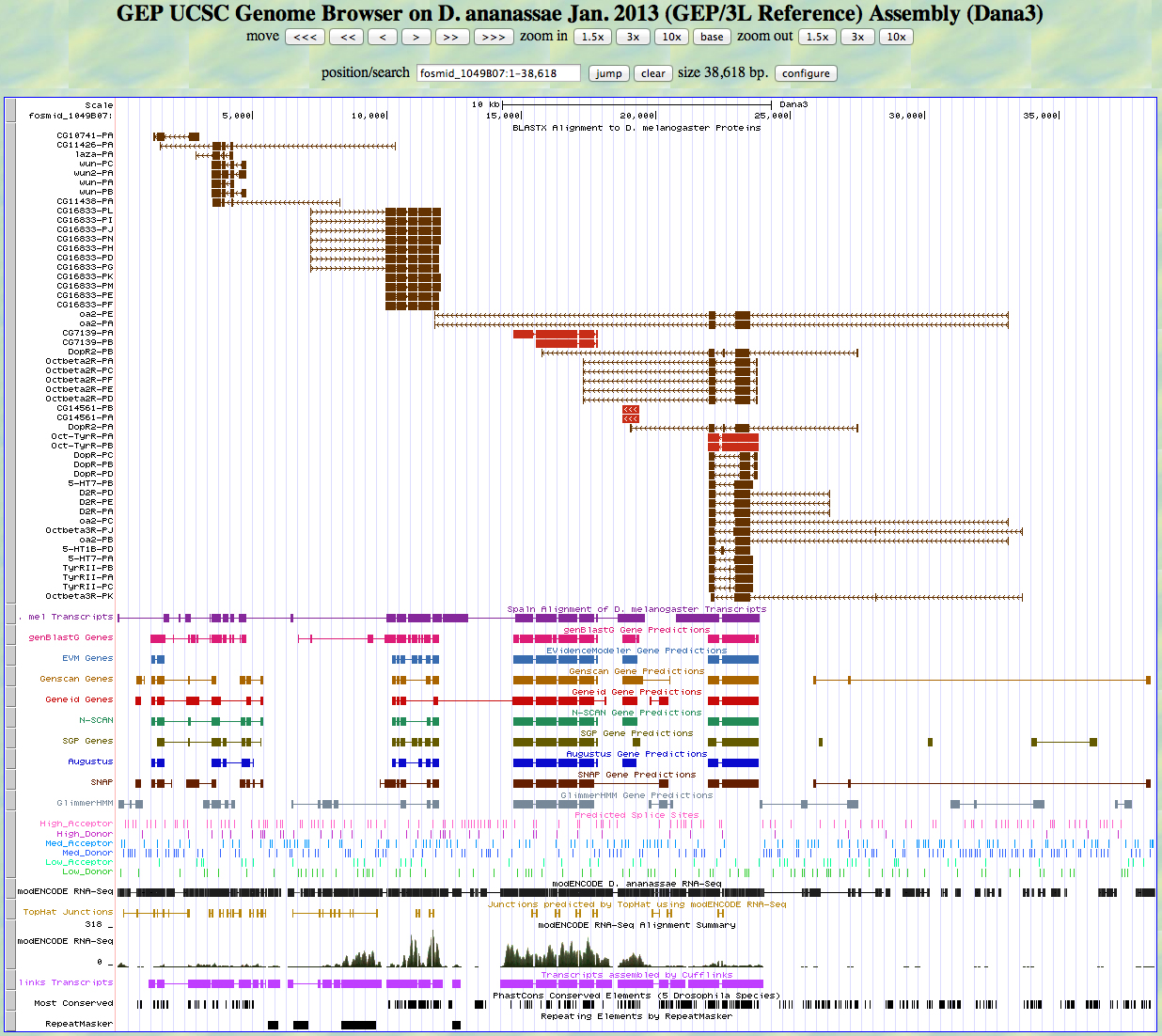
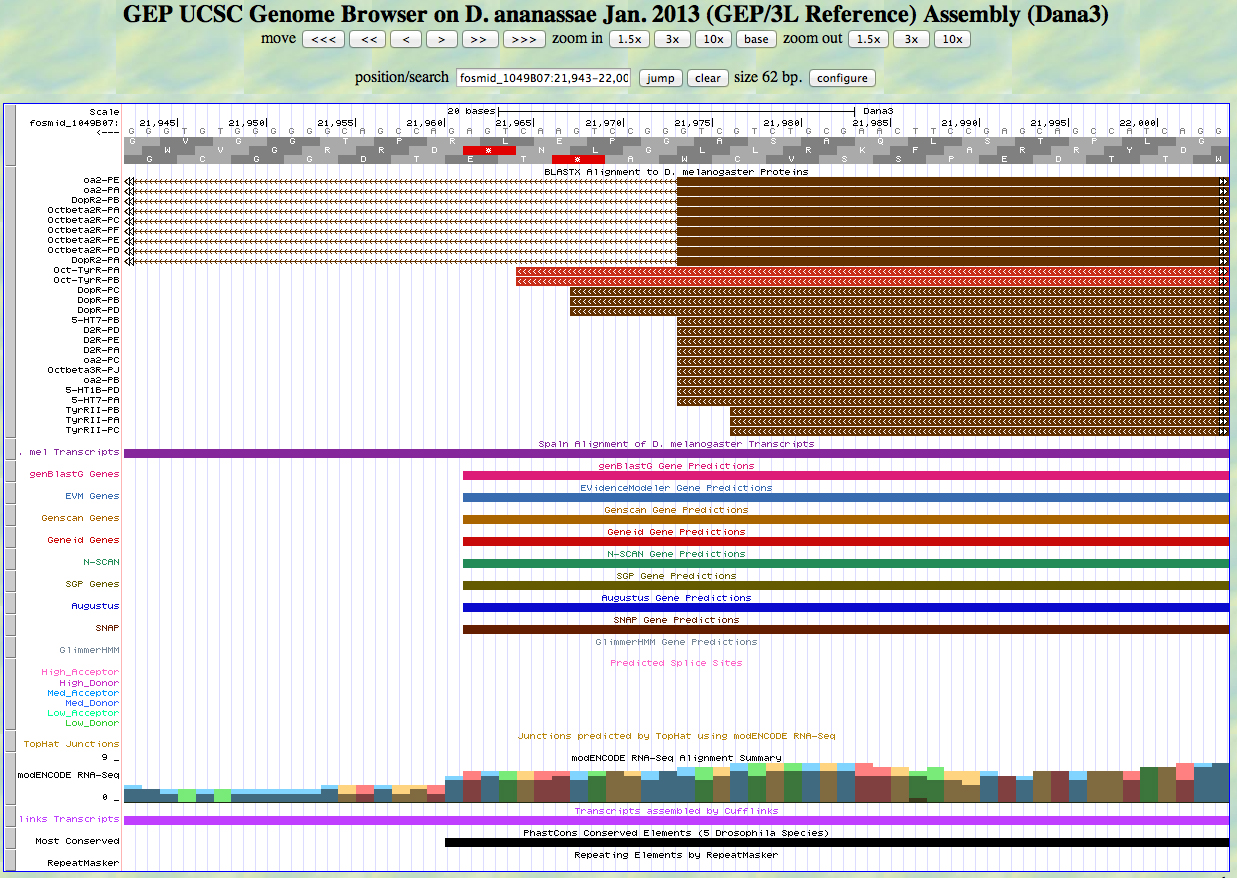
Here is the GEP UCSC genome browser view of D. ananassae fosmid 1049B07 from the 3L control region.

We would like to show how to annotate genes on the minus strand in this example. Notice the cluster of aligned D. melanogaster proteins on the right. The direction of transcription is from right to left in this view (note the arrowheads in the introns). This means that this gene is on the minus strand.
Next we carry out a BLASTX search of the nr database of D. melanogaster proteins using the six-frame translation of D. ananassae fosmid 1049B07 as the query sequence. The top hits are shown below.
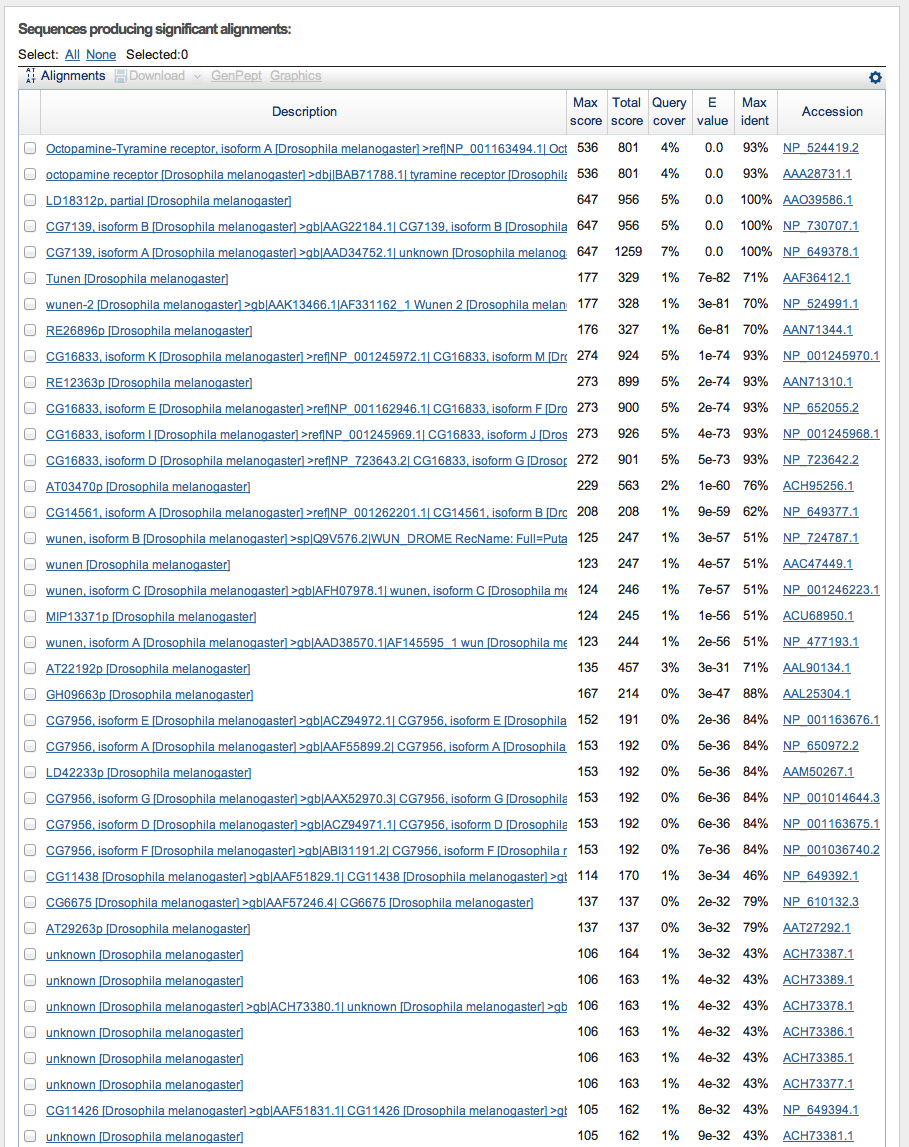
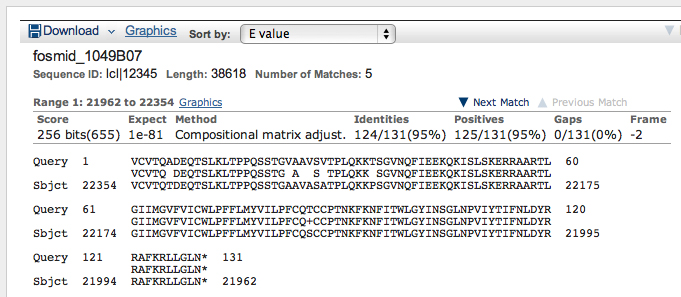
The top hit is the Octopamine-Tyramine receptor. The alignment is shown below.

Notice that the alignment is in two segments, to frame -3 and -2.
We go to FlyBase to discover the gene symbol. A search for "octopamine" gives the results shown below.
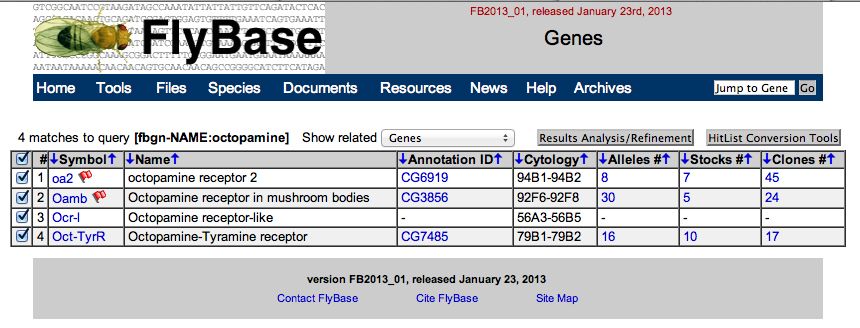
Only one gene, Oct-TyrR, is in the 79B region.
We go to the Gene Record Finder at GEP to retrieve the transcript and CDS information for this gene. The results are shown below.
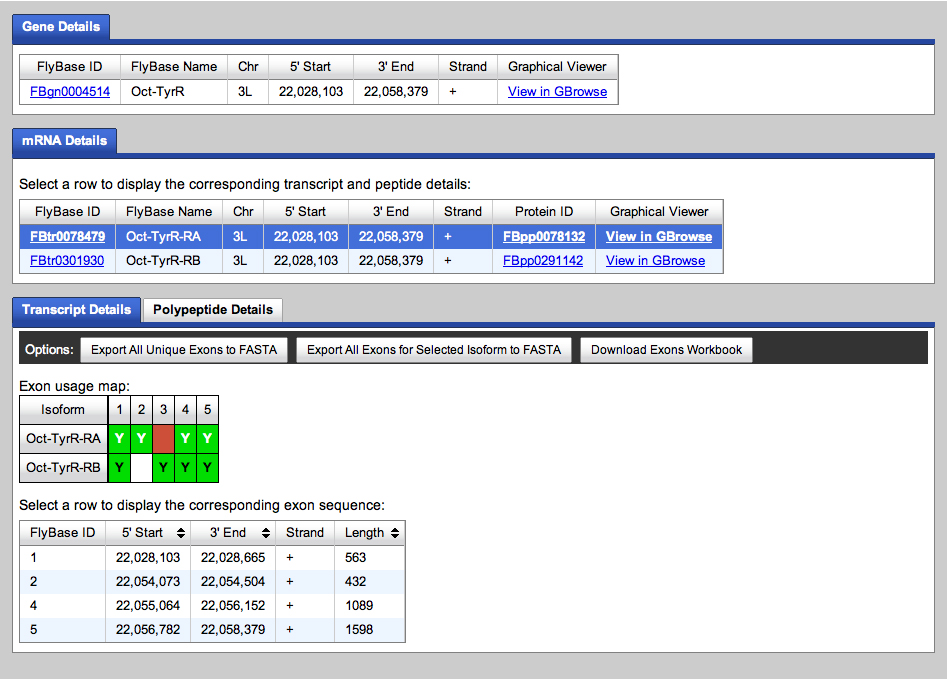
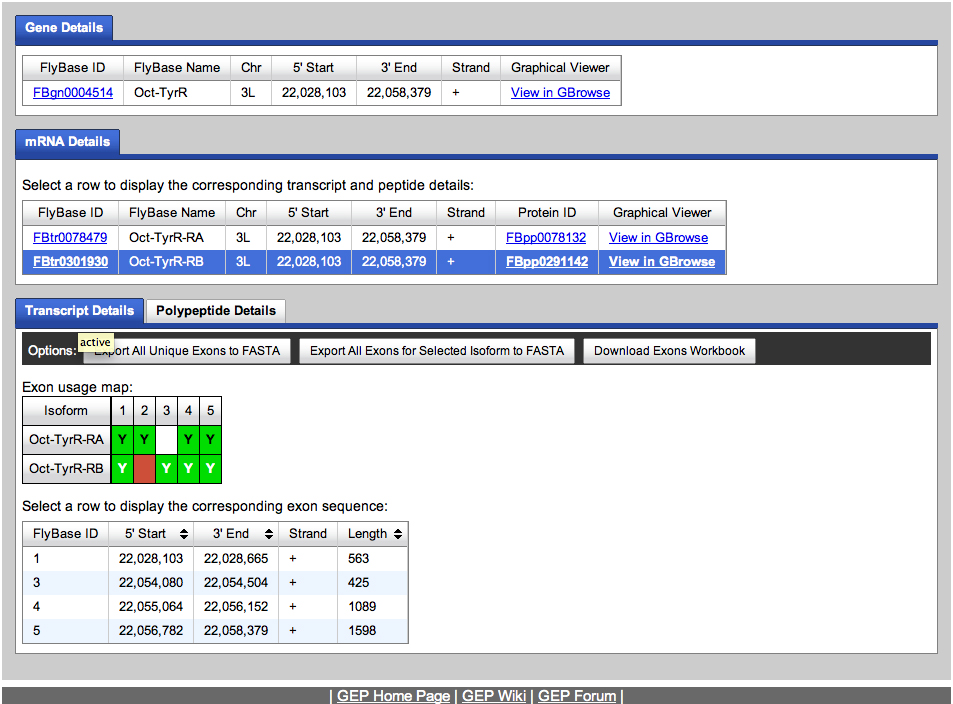
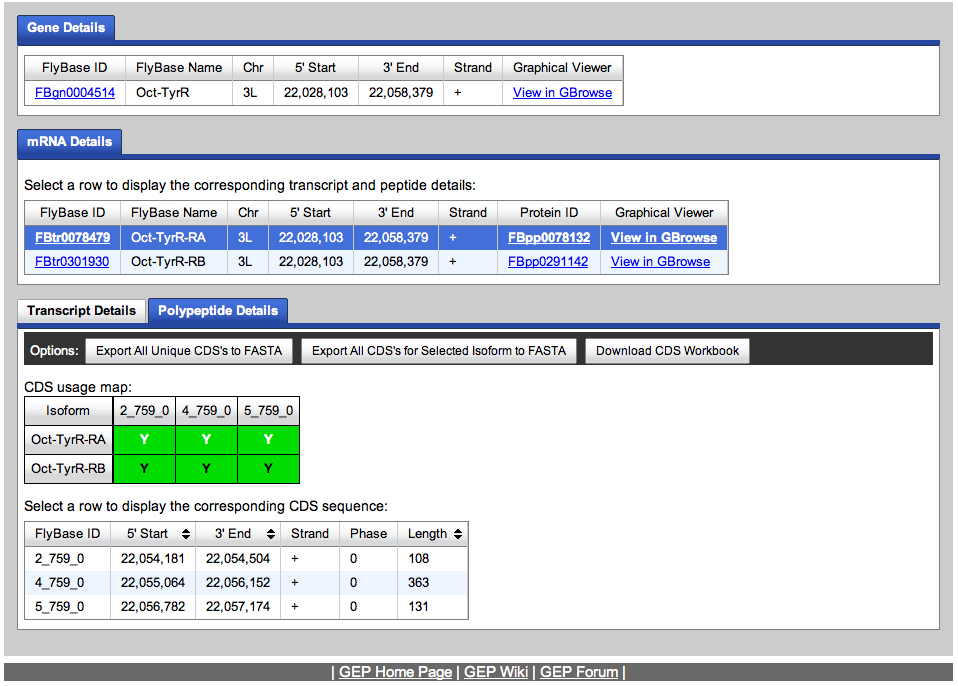
We retrieve the transcript and CDS data.
We will use the transcript data to begin annotating the Oct-TyrR gene on the fosmid.
We carry out five bl2seq BLASTN alignments of the D. ananassae fosmid 1049B07 (the subject sequence) and individual D. melanogaster exon sequences from the transcript data.
>Oct-TyrR:1 CGAATTGGCGTTGGGTGTGGACGCGAGTGGCGGAAGCAGGCGGCGTTGAT ATATACGAGCTCTTCCATCTTTCGTGATGCGGTATTAGAAGCTGGCAGCT CAGAGATTCCCGCAAAGTTTAAGTGACAATTTGCCAGCCAACAACAACTT TCCGACGCAGGCAACGCAAATTGAATTCGATTCGATTCGATTGTCTCTCG ATCTTCATCAATTCATTACGCACAGGAAAAGAGGGCGAACCGTAAAGTTC TGGTGAAAAAGTTTCCTGGGCTCCGTTGGCGTGGCAAAGCGACCGAAACC AAAACGAAATTTTGAAAATGAGCTTTGCTAACGACGGGCCAAACCAATTA ACAGAATTCGTTCTTGTGTAATAAATAAATTGCCAACAATTATAACTTGC AGTCCACTCAGGCATATTCAAATGAAATGTGCCACAAAATGTTTACGGTC ATTGCAACTCAAAGCGACAGACCATAGACGAGGTGCAAGGTGTTGTGGCA GTTGCAGAAAAACTAAAAGAAAGCCGTAAGGCTTGACCAAAAATTAATAA CTGATAAAAGCAG
Results: No significant match.
>Oct-TyrR:2 AAATAAGTCAAAGAAGTCGGGGAAATCGCACTCAACGTCCGCCTTTCCAC CAAGACGCATGTAAACGCAACCGGAGCCCAAAGAAGGCAAGTGGCAGGGC AGGGAAAGATGCCATCGGCAGATCAGATCCTGTTTGTAAATGTCACCACA ACGGTGGCGGCGGCGGCTCTAACCGCTGCGGCCGCCGTCAGCACCACAAA GTCCGGAAGCGGCAACGCCGCACGGGGCTACACGGATTCGGATGACGATG CGGGCATGGGAACGGAGGCGGTGGCTAACATATCCGGCTCGCTGGTGGAG GGCCTGACCACCGTTACCGCGGCATTGAGTACGGCTCAGGCGGACAAGGA CTCAGCGGGAGAATGCGAAGGAGCTGTGGAGGAGCTGCATGCCAGCATCC TGGGCCTCCAGCTGGCTGTGCCGGAGTGGGAG
Results are shown below.
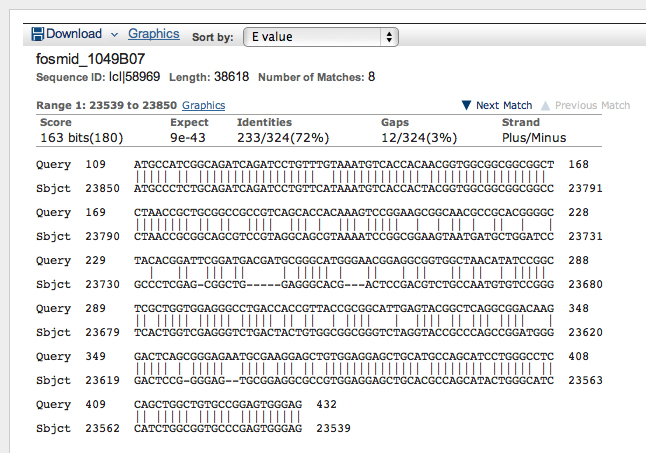
>Oct-TyrR:3 TCAAAGAAGTCGGGGAAATCGCACTCAACGTCCGCCTTTCCACCAAGACG CATGTAAACGCAACCGGAGCCCAAAGAAGGCAAGTGGCAGGGCAGGGAAA GATGCCATCGGCAGATCAGATCCTGTTTGTAAATGTCACCACAACGGTGG CGGCGGCGGCTCTAACCGCTGCGGCCGCCGTCAGCACCACAAAGTCCGGA AGCGGCAACGCCGCACGGGGCTACACGGATTCGGATGACGATGCGGGCAT GGGAACGGAGGCGGTGGCTAACATATCCGGCTCGCTGGTGGAGGGCCTGA CCACCGTTACCGCGGCATTGAGTACGGCTCAGGCGGACAAGGACTCAGCG GGAGAATGCGAAGGAGCTGTGGAGGAGCTGCATGCCAGCATCCTGGGCCT CCAGCTGGCTGTGCCGGAGTGGGAG
Results are shown below.
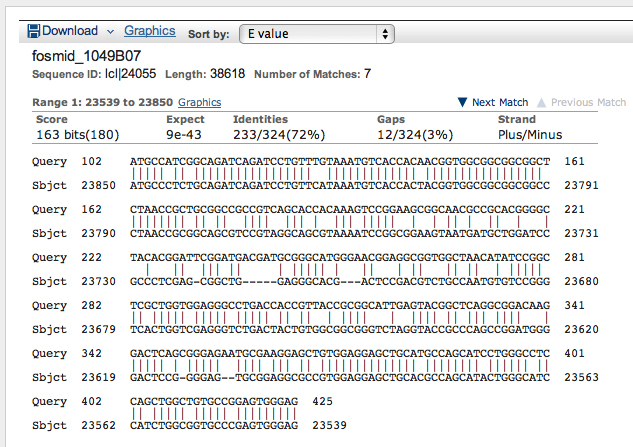
>Oct-TyrR:4 GCCCTTCTCACCGCCCTGGTTCTCTCGGTCATTATCGTGCTGACCATCAT CGGGAACATCCTGGTGATTCTGAGTGTGTTCACCTACAAGCCGCTGCGCA TCGTCCAGAACTTCTTCATAGTTTCGCTGGCGGTGGCCGATCTCACGGTG GCCCTTCTGGTGCTGCCCTTCAACGTGGCTTACTCGATCCTGGGGCGCTG GGAGTTCGGCATCCACCTGTGCAAGCTGTGGCTCACCTGCGACGTGCTGT GCTGCACTAGCTCCATCCTGAACCTGTGTGCCATAGCCCTCGACCGGTAC TGGGCCATTACGGACCCCATCAACTATGCCCAGAAGAGGACCGTTGGTCG CGTCCTGCTCCTCATCTCCGGGGTGTGGCTACTTTCGCTGCTGATAAGTA GTCCGCCGTTGATCGGCTGGAACGACTGGCCGGACGAGTTCACAAGCGCC ACGCCCTGCGAGCTGACCTCGCAGCGAGGCTACGTGATCTACTCCTCGCT GGGCTCCTTCTTTATTCCGCTGGCCATCATGACGATCGTCTACATCGAGA TCTTCGTGGCCACGCGGCGCCGCCTAAGGGAGCGAGCCAGGGCCAACAAG CTTAACACGATCGCTCTGAAGTCCACTGAGCTCGAGCCGATGGCAAACTC CTCGCCCGTCGCCGCCTCCAACTCCGGCTCCAAGTCGCGTCTCCTAGCCA GCTGGCTTTGCTGCGGCCGGGATCGGGCCCAGTTCGCCACGCCTATGATC CAGAACGACCAGGAGAGCATCAGCAGTGAAACCCACCAGCCGCAGGATTC CTCCAAAGCGGGTCCCCATGGCAACAGCGATCCCCAACAGCAGCACGTGG TCGTGCTGGTCAAGAAGTCGCGTCGCGCCAAGACCAAGGACTCCATTAAG CACGGCAAGACCCGTGGTGGCCGCAAGTCGCAGTCCTCGTCCACATGCGA GCCCCACGGCGAGCAACAGCTCTTACCCGCCGGCGGGGATGGCGGTAGCT GCCAGCCCGGCGGAGGCCACTCTGGAGGCGGAAAGTCGGACGCCGAGATC AGCACGGAGAGCGGGAGCGATCCCAAAGGTTGCATACAG
Results are shown below.

>Oct-TyrR:5 GTCTGCGTGACTCAGGCGGACGAGCAAACGTCCCTAAAGCTGACCCCGCC GCAATCCTCGACGGGAGTCGCTGCCGTTTCTGTCACTCCGTTGCAGAAGA AGACTAGTGGGGTTAACCAGTTCATTGAGGAGAAACAGAAGATCTCGCTT TCCAAGGAGCGGCGAGCGGCTCGCACCCTGGGCATCATCATGGGCGTGTT CGTCATCTGCTGGCTGCCCTTCTTCCTCATGTACGTCATTCTGCCCTTCT GCCAGACCTGCTGCCCCACGAACAAGTTCAAGAACTTCATCACCTGGCTG GGCTACATCAACTCGGGCCTGAATCCGGTCATCTACACCATCTTCAACCT GGACTACCGCCGGGCCTTCAAGCGACTTCTGGGCCTGAATTGAGGCTGGC TGGCGGGGGCGGGTGGAAGATATAAACCGGGCCAATCATGGTTCGAGCGG GAGAGCGTAACTCAAAGTTTGTGCCAAACTTAAATGGTCGTGTATGCGTT CAGCGGAGATCTCAGTCTATGTACAGTTGGACCCCCAGTTGATGAACTTC CGAGTTCAACTTCTCTAACACATATATACTTTCAAATGCGTTCTTGGTGA ACTCATTTTGAAGAAGTGGATGAATTTGGTAAAGTGTAATAGATTGAATA TAATTTTTAATGTTTAACGTTTCGGCAAAGTGAAAAGCCCCCACATTGGA AAGTCAAAGATGAGACTCGAGTGTATATATAGTTTCAAACTAAGTTATTA TTTCTAGCCGTAATTAAAATACTTTCATTTAGTTTTGAACATTTTTTTAA TATATTGTTGTTTGGAATCGATTGAGATGTACCACCACATTAAGCGTAGA TTGTTCAATACTCATACTAAAATGGGTTGTGCTGCGATTAAAGTGAGGAT GTTGCCTCAAGGCACAGCTACTAGGAAAATCATAAAAATTACATGGTAAA GAATTATACATGCATTATACTCCAGCTAAGTGGCATCCCAAACGAGAATA GCATCAAATTGAATTTAATACAATTAAATTAAATGTTTAGGCACAAAGAA TTGTGGCAACTTTCGTGTTTCACCCTAAGCGTATGGATAACCAAAAAGGT GTTTGTTAAATTAAATCTGCGCTCAAGATATGTAAGCAACTACTAAGCTA AATAATAACTTCCAAGAGAGAAACGTTTTCTAGGCATTACTTTAACGATT TGTATTTATATGTACTTTAATTGTAGGTAAACGATAAACCACTATACCTA ATGTATACTTTCAAATACGCTTTGGACTATTTGTTAAATAATTTAACGAT TAATTGTTTTTATGGCATAGCAACTATTGTGTTGAGTGGGCAGCTTAAAG CTAGCACATCGAAACTTACTTAAGGTAGATAAATGTTTAACTGCACGTTA CGAAATGCAACAGAGTTGGCGAAAGGACGTAATTCAATGGATGTGTTAAC TCAAGTACATGCTATATCGTAAATGTATATCACAATTTATGTCTTTTAAC GACGATGTACGATAGTTTCACTAATTATATTGTTTAACGAGAAAGAGCGA GCAAAGCGTAAATGAAACAAATAAAAGACACATTCGAATTAAAGTTAT
Results are shown below.
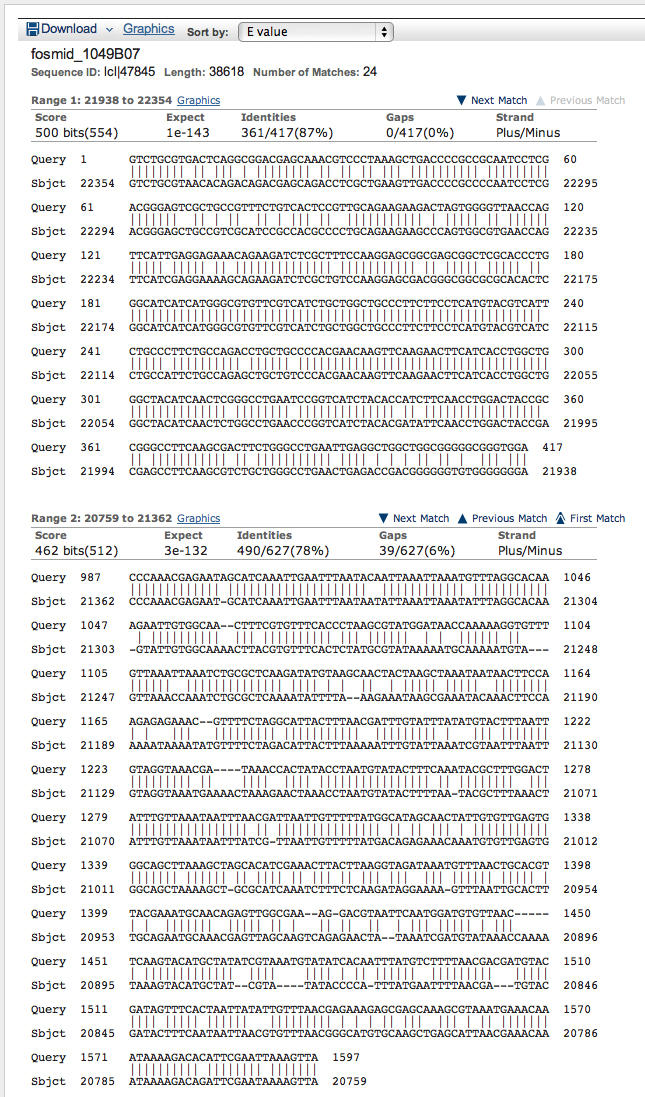
We will use the CDS data to begin annotating the Oct-TyrR gene on the fosmid.
We carry out three bl2seq TBLASTN alignments of the D. ananassae fosmid 1049B07 (the subject sequence) and individual D. melanogaster CDS segments (the query sequences) as shown below.
>Oct-TyrR:2_759_0 MPSADQILFVNVTTTVAAAALTAAAAVSTTKSGSGNAARGYTDSDDDAGM GTEAVANISGSLVEGLTTVTAALSTAQADKDSAGECEGAVEELHASILGL QLAVPEWE
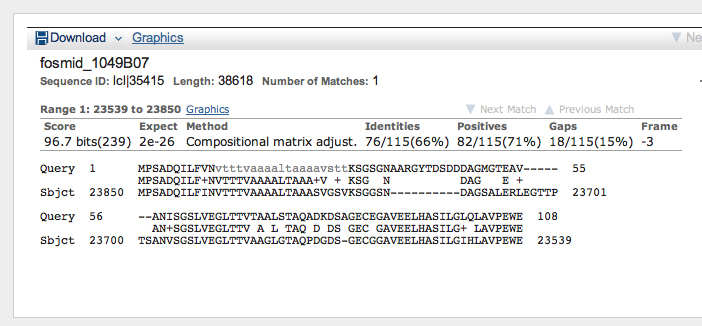
>Oct-TyrR:4_759_0 ALLTALVLSVIIVLTIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLTV ALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRY WAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANK LNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDRAQFATPMI QNDQESISSETHQPQDSSKAGPHGNSDPQQQHVVVLVKKSRRAKTKDSIK HGKTRGGRKSQSSSTCEPHGEQQLLPAGGDGGSCQPGGGHSGGGKSDAEI STESGSDPKGCIQ
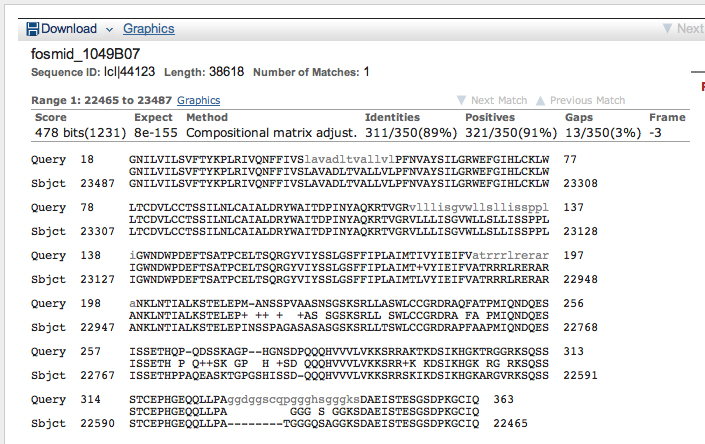
>Oct-TyrR:5_759_0 VCVTQADEQTSLKLTPPQSSTGVAAVSVTPLQKKTSGVNQFIEEKQKISL SKERRAARTLGIIMGVFVICWLPFFLMYVILPFCQTCCPTNKFKNFITWL GYINSGLNPVIYTIFNLDYRRAFKRLLGLN*
This gives us the information shown below.
| Dmel Exon | Dana Start | Dana End |
| Oct-TyrR:1 | N/A | N/A |
| Oct-TyrR:2 | 23850 | 23539 |
| Oct-TyrR:3 | 23850 | 23539 |
| Oct-TyrR:4 | 23538 | 22465 |
| Oct-TyrR:5 A | 22354 | 21938 |
| Oct-TyrR:5 B | 21362 | 20759 |
| Dmel Exon | Dana Start | Dana End | Frame |
| CDS 2 Oct-TyrR:2_759_0 |
23850 | 23539 | -3 |
| CDS 4 Oct-TyrR:4_759_0 |
23487 | 22465 | -3 |
| CDS 5 Oct-TyrR:5_759_0 |
22354 | 21962 | -2 |
Notice that the coordinates of the start of the coding segments on the fosmid are larger than the coordinates of the end of the coding segments. When we read the minus strand, we move from right to left, that is, from high coordinates to low coordinates.
There is an unusual feature here. Notice that Exon 3 and Exon 4 from the D. melanogaster transcript align to a single continuous sequence: the alignment of D. melanogaster Exon 3 ends at 23539 on the fosmid; the alignment of D. melanogaster Exon 4 begins at 23538. There is no intron separating Exons 3 and 4 in D. ananassae.
Now we go back to the GEP UCSC genome browser to locate the beginning and end of the first exon. Part of the view is shown below.
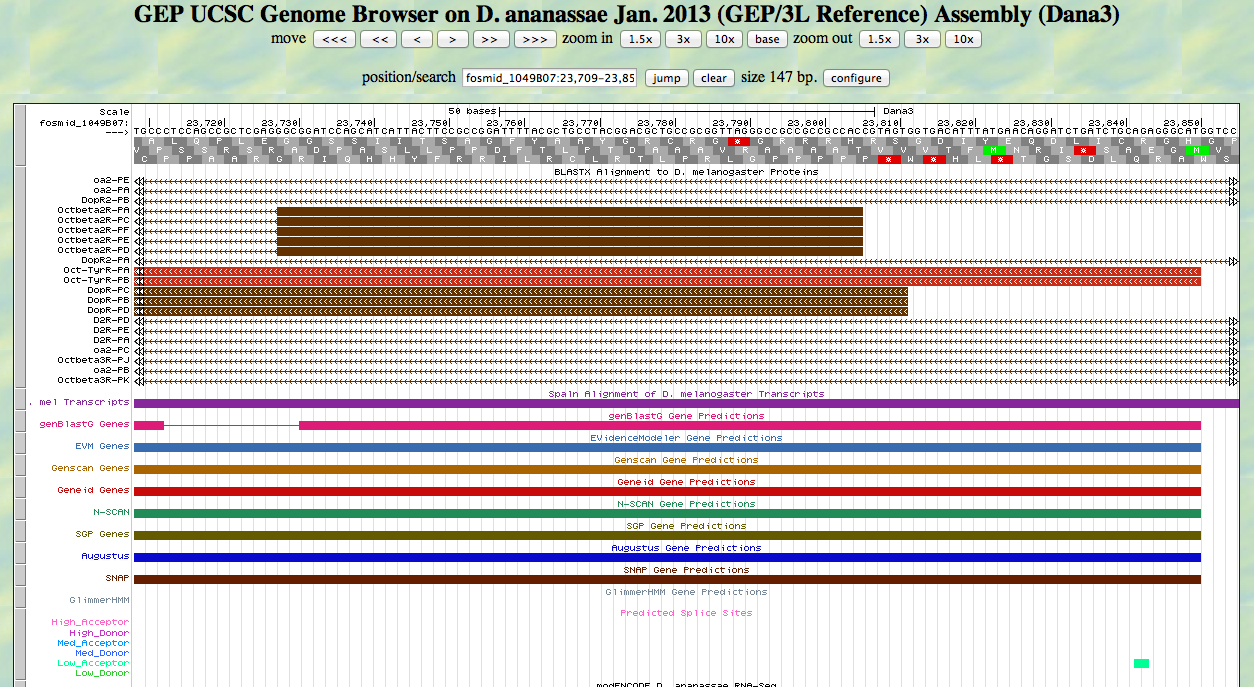
We would like to locate the ATG that begins at 23,850 according to our alignment. Notice in the screenshot above that the D. melanogaster Oct-TyrR isoforms align to this position.
We use scroll and zoom to move in closer, as shown below.
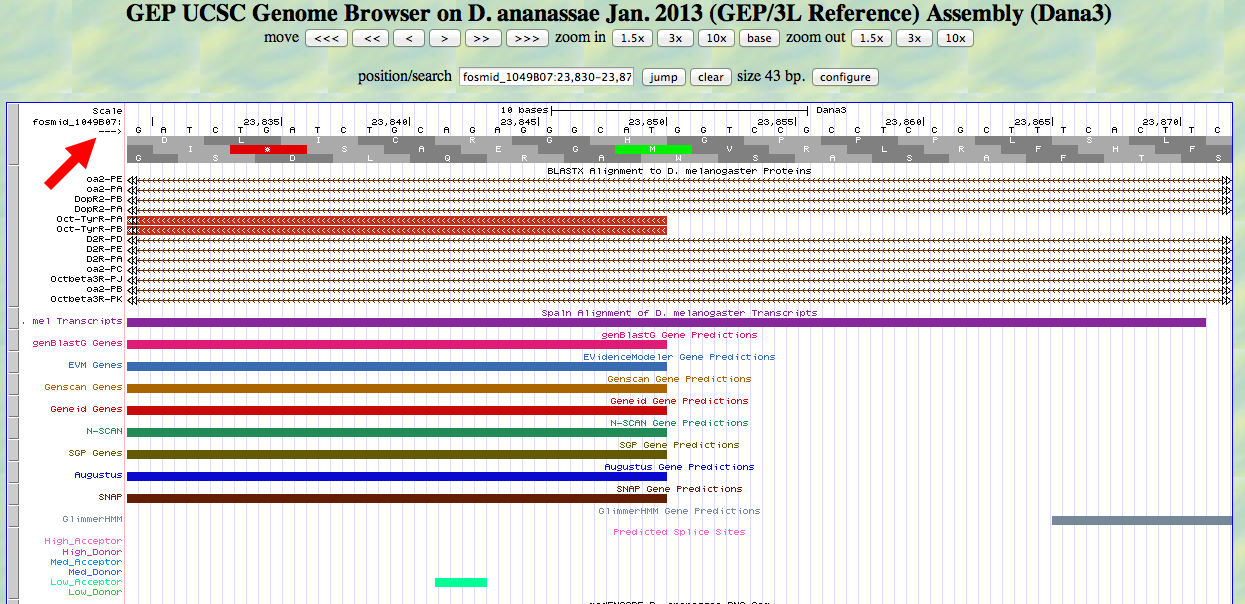
At first glance, this looks OK. We have an ATG and a methionine at about the right position. However, we know that the alignment is to the reading frame -3. We are viewing an ATG in reading frame +2. We can read the ATG in frame +2 from left to right, but the CDS for this gene is read from right to left. Note the red arrow pointing to the small arrow in the upper left. Clicking the small arrow changes the display as shown below.
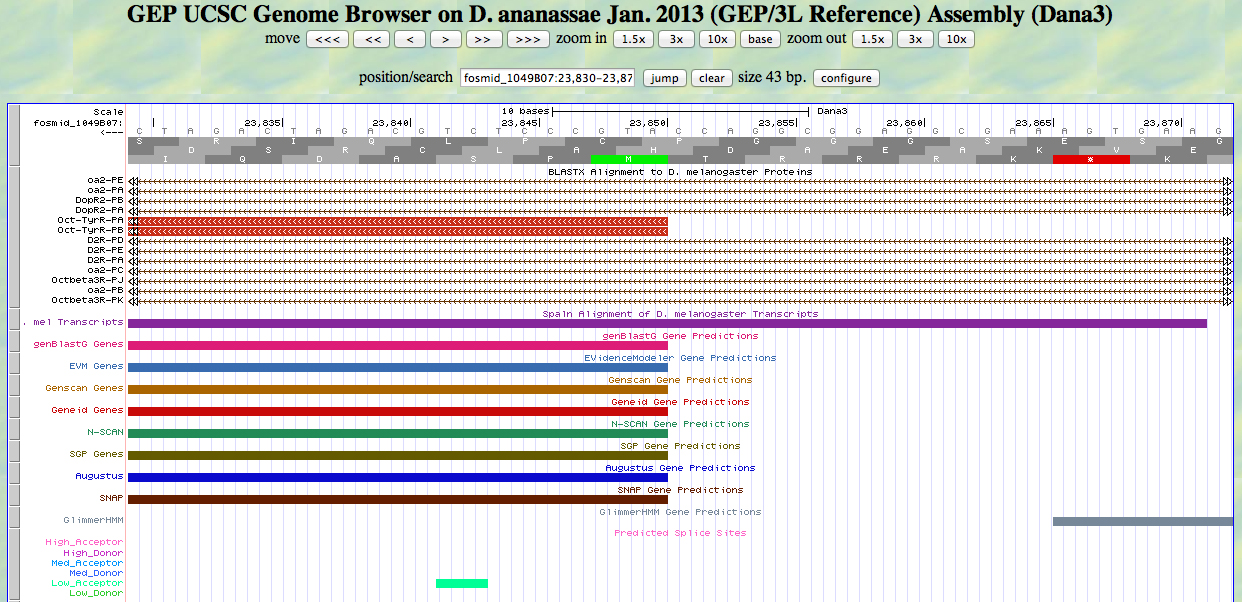
Now we see a methionine in frame -3, and we can read the ATG from right to left. The gray text for the DNA sequence (as opposed to the black text in the previous screenshot) shows that we are reading the minus strand.
We confirm that the position of the first base of the ATG is 23850.
While the first segment of our D. melanogaster CDS alignment ends at 23539, reading frame -3 is open all the way through the end of the second aligning segment at 22465. Let's see if we can find the end of the second aligning segment. We use scroll and zoom to arrive at the view shown below.
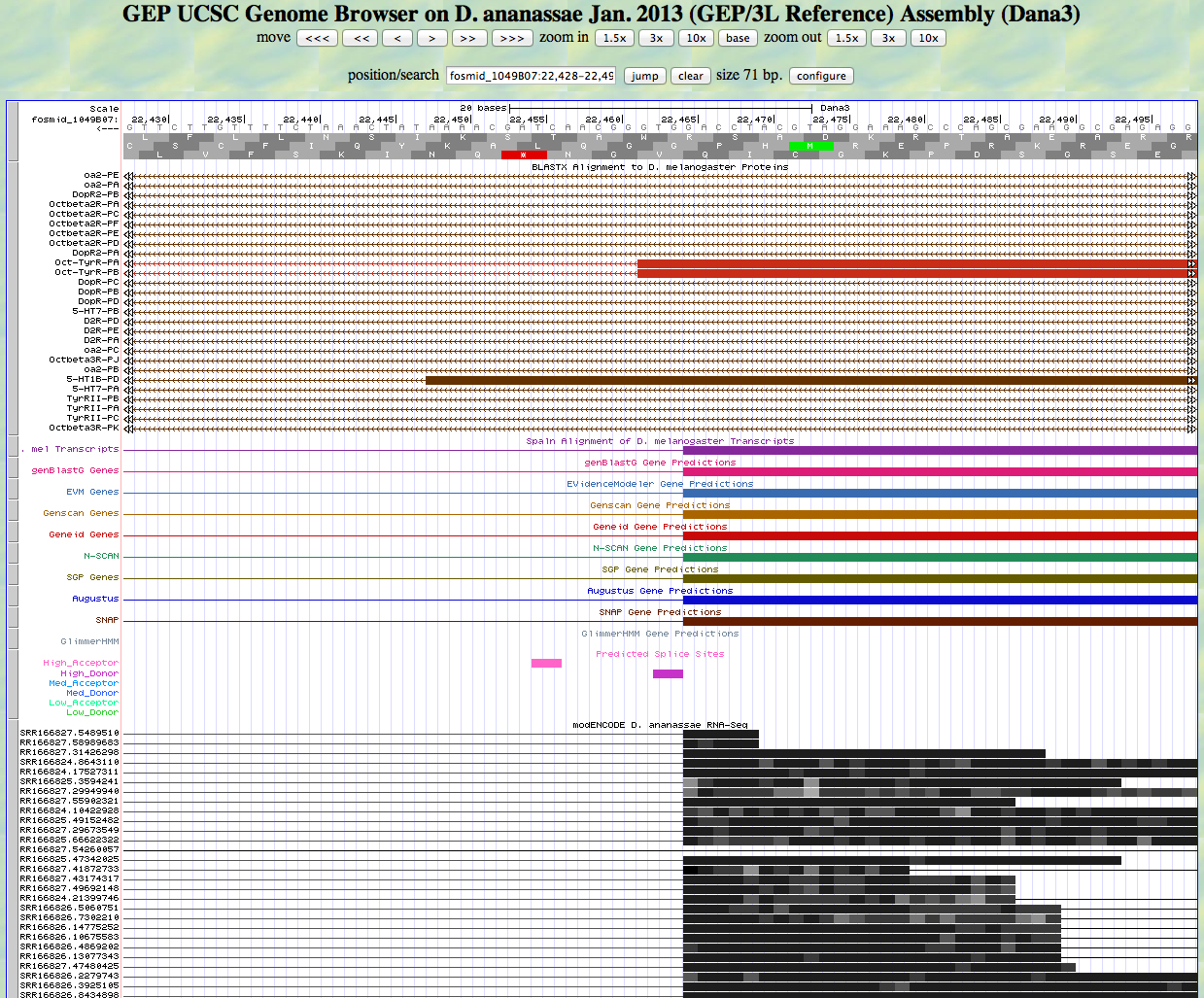
There is a high donor site after 22465. We can read the GT from right to left. The RNA-Seq tracks support a splice here. All of the gene prediction programs call a splice here also. Using frame -3, we see that the next exon is phase 0.
We zoom out and turn off the detailed RNA-Seq track to get the view below.

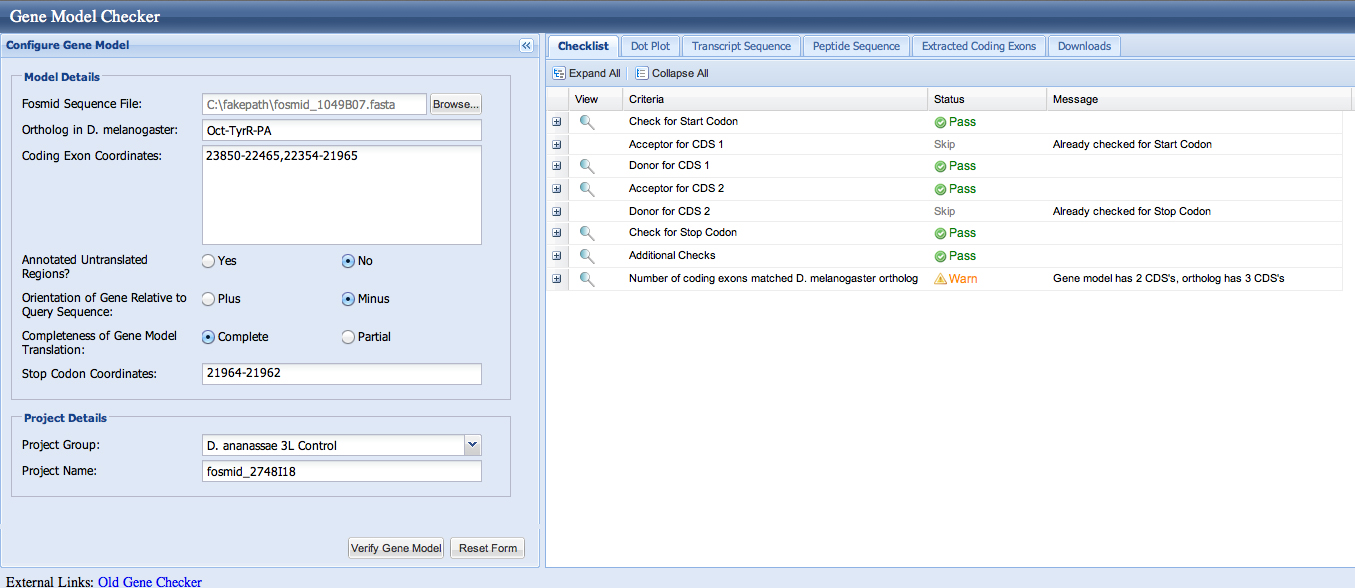
The modENCODE RNA-Seq data support the idea that the first exon runs from 23850-22465. We can use the Gene Model Checker to verify Exon 4, as shown below.
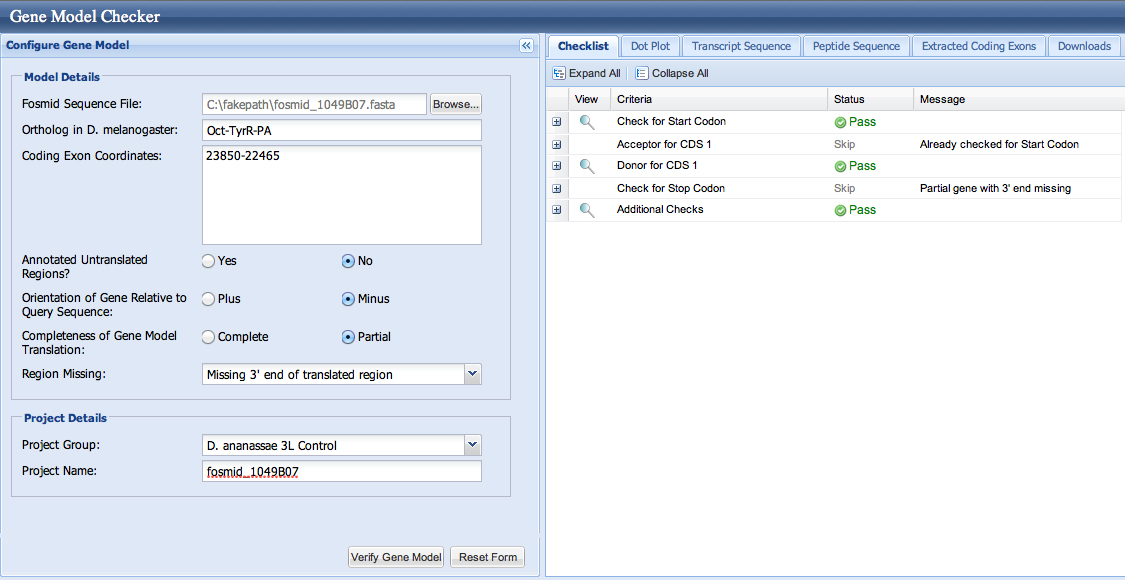
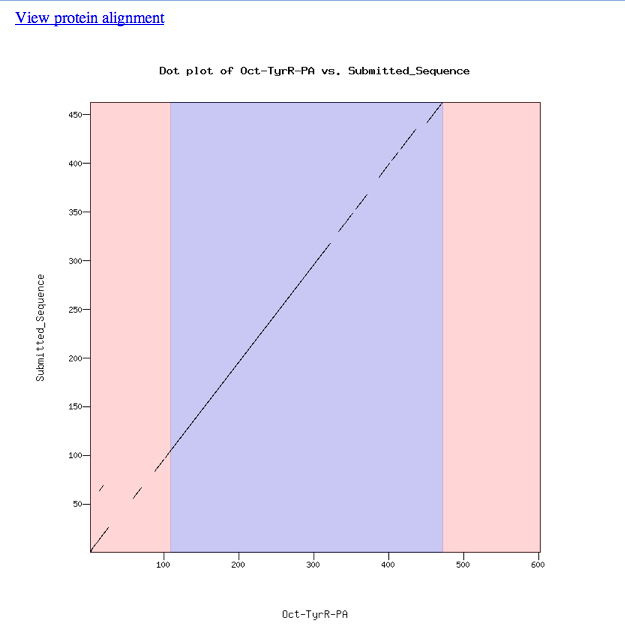
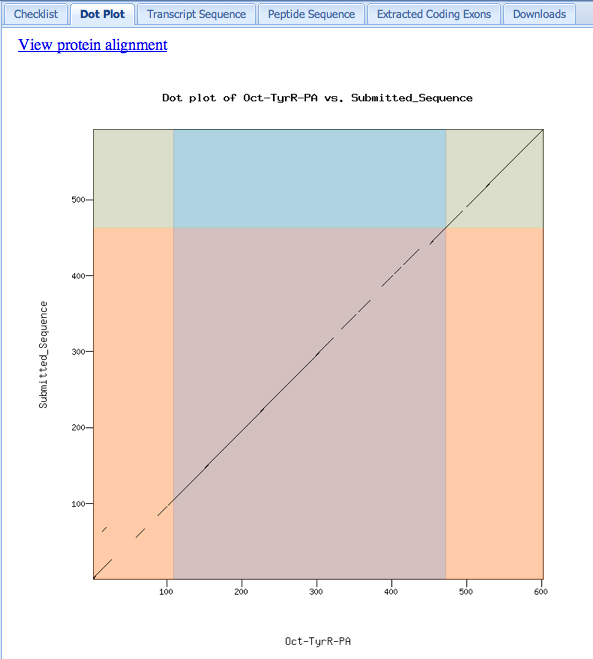
Our model for Exon 4 looks good. The dotplot is shown below. Notice that the dotplot is color-coded to conform to the D. melanogaster exons. Our model, which merges the first and second exons, aligns perfectly with the D. melanogaster protein. This appears to be one of those rare instances when we have lost an intron in D. ananassae, or gained one in D. melanogaster. The alignment is offset a bit near the end of our first exon. This seems to be a loss of some amino acids in D. ananassae relative to D. melanogaster at this position.
Here is the last exon.
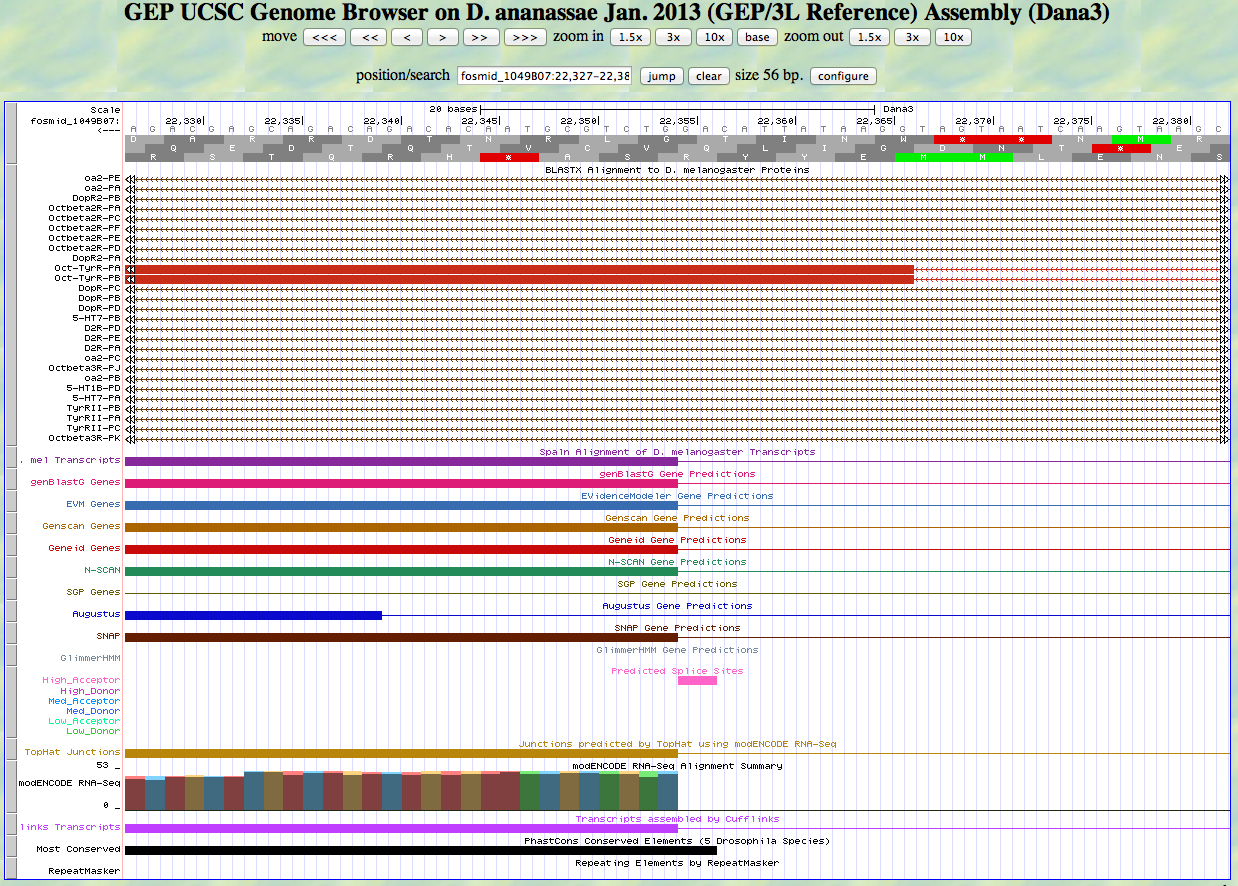
There is a high acceptor AG that ends at 22355. Most of the gene predictors call a splice here, which is supported by the RNA-Seq data. The exon begins at 22354. Using frame -2, we see that this exon is phase 0 as predicted.
We move to the end of the second exon, as shown below.
The end of the coding sequence in frame -2 is 21965. The protein alignment ends here exactly. The stop is 21964-21962. The gene predictors call the stop here.
Here is a check using the Gene Model Checker. We are warned that the number of coding exons does not match the D. melanogaster ortholog. The dot plot looks really good, giving us confidence in the model, but it is necessary to look more carefully into this.
Here is the D. melanogaster genomic sequence for the region including Oct-TyrR from FlyBase.
>3L:22053241,22058240 agttcagtttatgagcgggttattttgattaatgttccttcaactggtgtcaactggtag gcggacaaaaaggagcccgatagcaaaattcagcaaagctggtaaggacttagtggatag tgaagtgtcaggcggaagtgaaaagagcagagcaacaattattcgatgaccacagaaggg gagtggggagcgtattgacttagggcaaaaagtgatatccctgaacatcttagaaccctc tttccttatcgacacacatttacttaggaccatctttttgacccaattcgttctgacatt tttaattacttttagataattaatcccaagttcagataattcgagaagatttgtctgtaa aattgttagggaccttcccaccgtgattcctttaggttttcttttgatggtcccttctat cgaatgtcctttgggcttattgacaagctgtaaataatttggcatgacatgatcctgata gccttgtgcggtaactttgtccaccctgatccgtaagtctgaactccttctaaacgcaat ttgtaaacctataaaaaaagaaaaaagcagctctcatattctttcgtaattatgtgaatg caagttaaaagtttaagccgattgagccacaattttgtctttccaattgatattagcaaa cagctatggtatagttattttaatacaatgccatctccatacatggttttttactctctg cgtacaatacagttaagttaagtaagccagaaaatttcaacgaaaattcccgcctcccaa cgaagtgttgaagtcacaataatttgctttcctctcaattttcctttcgcagaaataagt caaagaagtcggggaaatcgcactcaacgtccgcctttccaccaagacgcatgtaaacgc aaccggagcccaaagaaggcaagtggcagggcagggaaagatgccatcggcagatcagat cctgtttgtaaatgtcaccacaacggtggcggcggcggctctaaccgctgcggccgccgt cagcaccacaaagtccggaagcggcaacgccgcacggggctacacggattcggatgacga tgcgggcatgggaacggaggcggtggctaacatatccggctcgctggtggagggcctgac caccgttaccgcggcattgagtacggctcaggcggacaaggactcagcgggagaatgcga aggagctgtggaggagctgcatgccagcatcctgggcctccagctggctgtgccggagtg ggaggtaataagcacaatgatagtatttcatttcattactaataataacggatagcttct gcctttttgttttttattaacgtttgacagcaaaaataaaatattcacatgcaaacctta ccttttaaaaaattgcgttataaatccccttttacttttgaccattattttgatcaagta tttgtatacccaaccgcctgtttgttcgtacttctgtctgtccgtccgtatatctgtccg tatgaacagaaactataaaagaatgtacaacaggaagttgagattaagcatattggtttt agttaacttttacgcagtgcaaccaaaatttaaaaaaatgttataattttaatagattct atcatttgtcttgccaatttctattgaaagttcgcctgccacaaatttcacttatattaa gtgtgaccgtcaccacgtagtgcaagtgaccctccctagaccaccaaactggtagtttct actctttcaataagtctaaataccagtagatcttatttatgttggtttaaagctctccct aacttcttttctattttaactaggcccttctcaccgccctggttctctcggtcattatcg tgctgaccatcatcgggaacatcctggtgattctgagtgtgttcacctacaagccgctgc gcatcgtccagaacttcttcatagtttcgctggcggtggccgatctcacggtggcccttc tggtgctgcccttcaacgtggcttactcgatcctggggcgctgggagttcggcatccacc tgtgcaagctgtggctcacctgcgacgtgctgtgctgcactagctccatcctgaacctgt gtgccatagccctcgaccggtactgggccattacggaccccatcaactatgcccagaaga ggaccgttggtcgcgtcctgctcctcatctccggggtgtggctactttcgctgctgataa gtagtccgccgttgatcggctggaacgactggccggacgagttcacaagcgccacgccct gcgagctgacctcgcagcgaggctacgtgatctactcctcgctgggctccttctttattc cgctggccatcatgacgatcgtctacatcgagatcttcgtggccacgcggcgccgcctaa gggagcgagccagggccaacaagcttaacacgatcgctctgaagtccactgagctcgagc cgatggcaaactcctcgcccgtcgccgcctccaactccggctccaagtcgcgtctcctag ccagctggctttgctgcggccgggatcgggcccagttcgccacgcctatgatccagaacg accaggagagcatcagcagtgaaacccaccagccgcaggattcctccaaagcgggtcccc atggcaacagcgatccccaacagcagcacgtggtcgtgctggtcaagaagtcgcgtcgcg ccaagaccaaggactccattaagcacggcaagacccgtggtggccgcaagtcgcagtcct cgtccacatgcgagccccacggcgagcaacagctcttacccgccggcggggatggcggta gctgccagcccggcggaggccactctggaggcggaaagtcggacgccgagatcagcacgg agagcgggagcgatcccaaaggttgcatacaggtaagactcttcgtaatcgtgaagatat tgcaagcccataattgttttacacatttattttaataaaaatatttgtaataaagggtct tgtgtttacaactgctaataagtctagataataaatacatatacatattatacccgttac ttatcgagtataagggtatactagattcgttgaaaagtatttaacaggccgactatataa agtatataaattcttgatcaggatcaaaagccgagtcgatctaggtatgtccgtctgtcc gattgtccgtatgaacgttgagatctcaggaaatataaaaactagaaggctcagacaaag acgcagcgcaactttgactcatgttgccacgcccacaaaccgcacaaaactgccccgccc aaacttttgaaaaacgtcttgatattttttcatatttttataagccttgtaaatttctat ctatttgccacgctagggtatctaatattcagggaaatcgacaaaaaaattaaaaatcta aagtgctcttttatttagagagccacatggaacaagcgatataagttgtataagttgtat gggagaaatggctttttccatgccacttgattaattaaacttttcctcacctcgattcca ggtctgcgtgactcaggcggacgagcaaacgtccctaaagctgaccccgccgcaatcctc gacgggagtcgctgccgtttctgtcactccgttgcagaagaagactagtggggttaacca gttcattgaggagaaacagaagatctcgctttccaaggagcggcgagcggctcgcaccct gggcatcatcatgggcgtgttcgtcatctgctggctgcccttcttcctcatgtacgtcat tctgcccttctgccagacctgctgccccacgaacaagttcaagaacttcatcacctggct gggctacatcaactcgggcctgaatccggtcatctacaccatcttcaacctggactaccg ccgggccttcaagcgacttctgggcctgaattgaggctggctggcgggggcgggtggaag atataaaccgggccaatcatggttcgagcgggagagcgtaactcaaagtttgtgccaaac ttaaatggtcgtgtatgcgttcagcggagatctcagtctatgtacagttggacccccagt tgatgaacttccgagttcaacttctctaacacatatatactttcaaatgcgttcttggtg aactcattttgaagaagtggatgaatttggtaaagtgtaatagattgaatataattttta atgtttaacgtttcggcaaagtgaaaagcccccacattggaaagtcaaagatgagactcg agtgtatatatagtttcaaactaagttattatttctagccgtaattaaaatactttcatt tagttttgaacatttttttaatatattgttgtttggaatcgattgagatgtaccaccaca ttaagcgtagattgttcaatactcatactaaaatgggttgtgctgcgattaaagtgagga tgttgcctcaaggcacagctactaggaaaatcataaaaattacatggtaaagaattatac atgcattatactccagctaagtggcatcccaaacgagaatagcatcaaattgaatttaat acaattaaattaaatgtttaggcacaaagaattgtggcaactttcgtgtttcaccctaag cgtatggataaccaaaaaggtgtttgttaaattaaatctgcgctcaagatatgtaagcaa ctactaagctaaataataacttccaagagagaaacgttttctaggcattactttaacgat ttgtatttatatgtactttaattgtaggtaaacgataaaccactatacctaatgtatact ttcaaatacgctttggactatttgttaaataatttaacgattaattgtttttatggcata gcaactattgtgttgagtgggcagcttaaagctagcacatcgaaacttacttaaggtaga taaatgtttaactgcacgttacgaaatgcaacagagttggcgaaaggacgtaattcaatg gatgtgttaactcaagtaca
Here is a bl2seq alignment of the sequences using BLASTN.
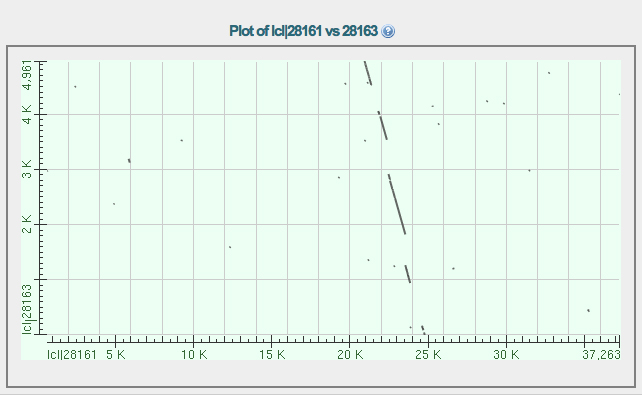
We see alignments to the D. ananassae fosmid in the range from around 21,000 - 24,000 as noted earlier in the BLAST alignments of the D. melanogaster transcript.
Using only part of the fosmid as the subject sequence gives us a closer look at the aligning segments, shown below.
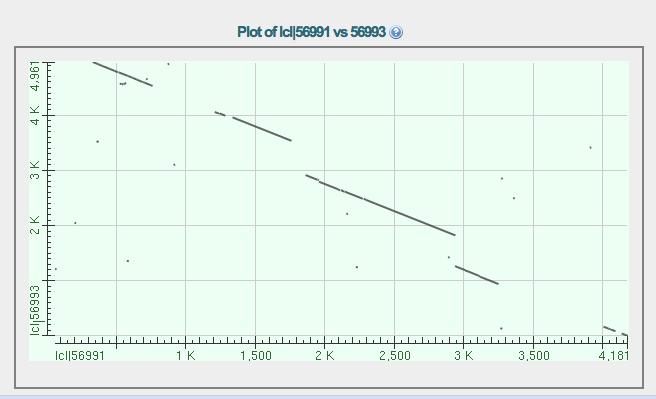
The alignments are in three gapped segments. Starting at the top left, the first two segments are in line. The middle segment is on a separate line. The gapped alignment in the lower right is on a third line. This shows that there are three segments of conserved sequence between D. melanogaster and D. ananassae, which is what we saw from the CDS alignments using BLAST.
Note the position on the X axis of the end of the second aligned segment and the beginning of the third. There is a substantial insertion in the D. melanogaster sequence relative to the D. ananassae sequence near 3,000 bp on the X axis. This is the missing intron in D. ananassae.
If this is correct, the position of this spot on the D. ananassae fosmid can be read from the first dotplot. It is located near 23,500 on the fosmid.
This position was noted earlier in our BLASTN alignment of the D. melanogaster transcript to the fosmid:
| Dmel Exon | Dana Start | Dana End |
| Oct-TyrR:1 | N/A | N/A |
| Oct-TyrR:2 | 23850 | 23539 |
| Oct-TyrR:3 | 23850 | 23539 |
| Oct-TyrR:4 | 23538 | 22465 |
| Oct-TyrR:5 A | 22354 | 21938 |
| Oct-TyrR:5 B | 21362 | 20759 |
Note that there is no gap in D. ananassae fosmid coordinates between the end of D. melanogaster Exon 3 and the beginning of Exon 4. We can use BLASTN to obtain the coordinates of the D. melanogaster exons on the assembly by using the D. melanogaster exons as query sequences and the D. melanogaster assembly as the subject sequence.
For this search, the query sequences are the exons from the D. melanogaster transcripts, the subject sequence is NCBI genomes restricted to Drosophila melanogaster, and the program is optimized for highly similar sequences (megablast). The results are shown below.
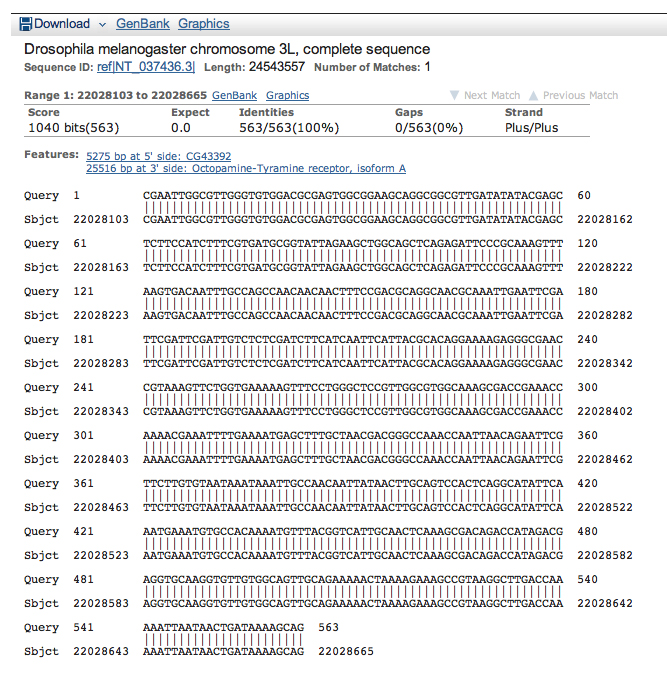
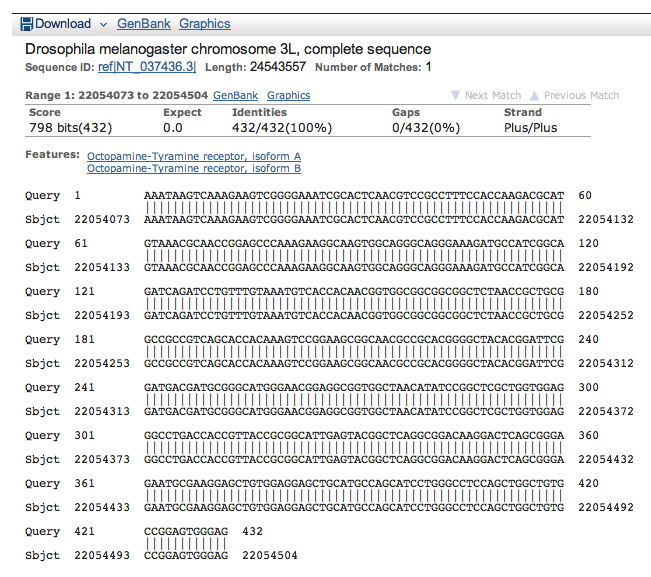
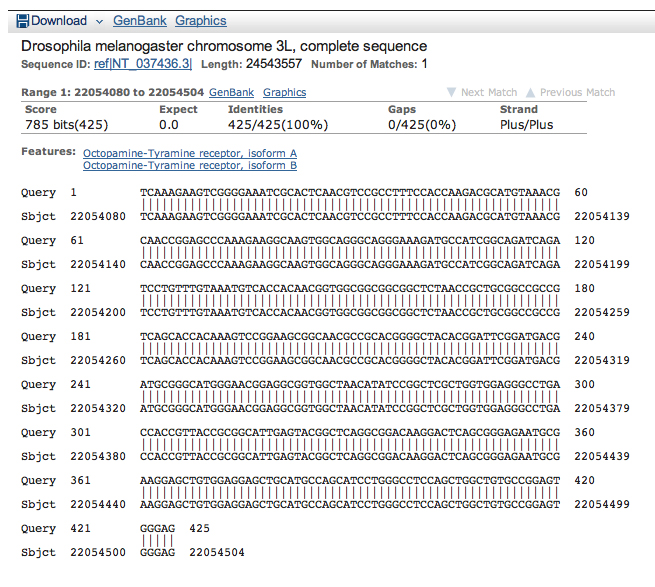


Tabulating the coordinates of the D. melanogaster alignments together with our earlier results gives us the results shown below.
| Dmel Exon | Dmel Start | Dmel End | Dana Start | Dana End |
| Oct-TyrR:1 | 22028103 | 22028665 | N/A | N/A |
| Oct-TyrR:2 | 22054073 | 22054504 | 23850 | 23539 |
| Oct-TyrR:3 | 22054080 | 22054504 | 23850 | 23539 |
| Oct-TyrR:4 | 22055064 | 22056152 | 23538 | 22465 |
| Oct-TyrR:5 A | 22056782 | 22058379 | 22354 | 21938 |
| Oct-TyrR:5 B | 22056782 | 22058379 | 21362 | 20759 |
These results show that:
1. D. melanogaster Exons 2 and 3 are nearly identical, with the same 3' end on both the D. melanogaster assembly and on the D. ananassae fosmid. The 5' ends differ by 7 bp on the D. melanogaster assembly, but the alignment to the D. ananassae fosmid ends at the same position for both exons.
2. There is an intron in D. melanogaster between the 3' end of Exon 2/3 and the 5' end of Exon 4. This intron is 560 bp in D. melanogaster, but is missing in D. ananassae.
3. The two segments of D. ananassae that align to Exon 5 of D. melanogaster are separated by a length of sequence that aligns poorly due to divergence, but is contiguous. This is best seen in the dotplots, where the first and second segments of aligning sequence in the upper left are on the same line.
The model is easier to understand in a drawing, shown below.
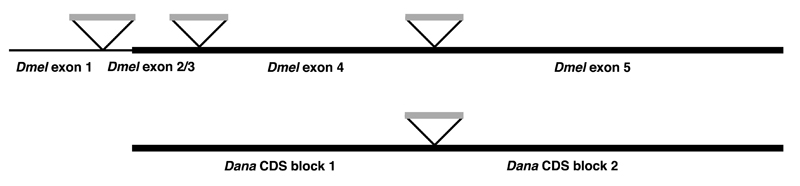
The drawing (introns not to scale) shows the D. melanogaster gene model on the top line and the D. ananassae gene model below. The D. melanogaster gene has a noncoding Exon 1, two very similar Exons 2 and 3 (shown as a single exon in the model) in which the CDS begins, a coding Exon 4, and the final coding Exon 5. The D. ananassae model has two blocks of CDS separated by a single intron.
It is interesting to investigate which Drosophila species contain the intron found in D. melanogaster but not in D. ananassae. To explore this question, I made a single protein sequence that consists of the CDS of Exons 2/3 and Exon 4 from D. melanogaster, shown below.
>Oct-TyrR:2-4_hybrid MPSADQILFVNVTTTVAAAALTAAAAVSTTKSGSGNAARGYTDSDDDAGM GTEAVANISGSLVEGLTTVTAALSTAQADKDSAGECEGAVEELHASILGL QLAVPEWE ALLTALVLSVIIVLTIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLTV ALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRY WAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANK LNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDRAQFATPMI QNDQESISSETHQPQDSSKAGPHGNSDPQQQHVVVLVKKSRRAKTKDSIK HGKTRGGRKSQSSSTCEPHGEQQLLPAGGDGGSCQPGGGHSGGGKSDAEI STESGSDPKGCIQ
This sequence was used to carry out a TBLASTN search of genus Drosophila (subgenus Sophophora and subgenus Drosophila) at FlyBase. The graphical summary is shown below.
If the most significant visual alignment breaks (with a dotted line) in the position near 100, it indicates the presence of the intron.

It is possible to divide the results into two categories: those with the intron (like D. melanogaster) and those without the intron (like D. ananassae). The results are summarized below.
| intron | no intron |
| D. melanogaster | D. ananassae |
| D. simulans | D. pseudoobscura |
| D. sechellia | D. persimilis |
| D. yakuba | D. willistoni |
| D. erecta | D. mojavensis |
| D. biarmipes | D. grimshawii |
| D. eugracilis | D. elegans |
| D. takahashii | D. rhopaloa |
| D. ficusphila | |
| D. kikkawai | |
| D. bipectinata | |
| D. miranda | |
| D. virilis |
Examination of the taxonomy of the Sequenced Species at FlyBase shows that D. ananassae, a member of the melanogaster group, is intronless. It branches off from the rest of the melanogaster subgroup, all of which have the intron (D. simulans, D. sechellia, D. melanogaster, D. yakuba, and D. erecta). Members of the more distantly related obscura group (D. pseudoobscura and D. virilis) lack the intron, as does the last remaining species in the Sophophora, D. willistoni. The three species in the distant Drosophila branch, D. mojavensis, D. virilis, and D. grimshawii, also lack the intron.
This suggests that gain of this intron in the melanogaster subgroup occurred after that group branched from D. ananassae around 20 million years ago, and that the ancestral form lacks the intron.