A multiple alignment is an alignment of more than two nucleotide or amino acid sequences. Constructing a multiple alignment is a useful way to compare different genes. A multiple alignment can help you to identify important conserved positions. You can also use a multiple alignment to construct a phylogenetic tree of related proteins.
In this example, we will use our gene model for the D. ananassae Oct-TyrR gene as the starting point. Here is the predicted peptide:
>Oct-TyrR-PA_peptide [Drosophila ananassae] MPSADQILFINVTTTVAAAALTAAASVGSVKSGGSNDAGSALERLEGTTPTSANVSGSLV EGLTTVAAGLGTAQPDGDSGECGGAVEELHASILGIHLAVPEWEALLTALVLSVIIVLTI IGNILVILSVFTYKPLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKL WLTCDVLCCTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPP LIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFIPLAIMTLVYIEIFVATRRRLRERA RANKLNTIALKSTELEPINSSPAGASASASGSKSRLLTSWLCCGRDRAPFAAPMIQNDQE SISSETHPPAQEASKTGPGSHISSDQQQHVVVLVKKSRRSKIKDSIKHGKARGVRKSQSS STCEPHGEQQLLPATGGGQSAGGKSDAEISTESGSDPKGCIQVCVTQTDEQTSLKLTPPQ SSTGAAVASATPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMY VILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
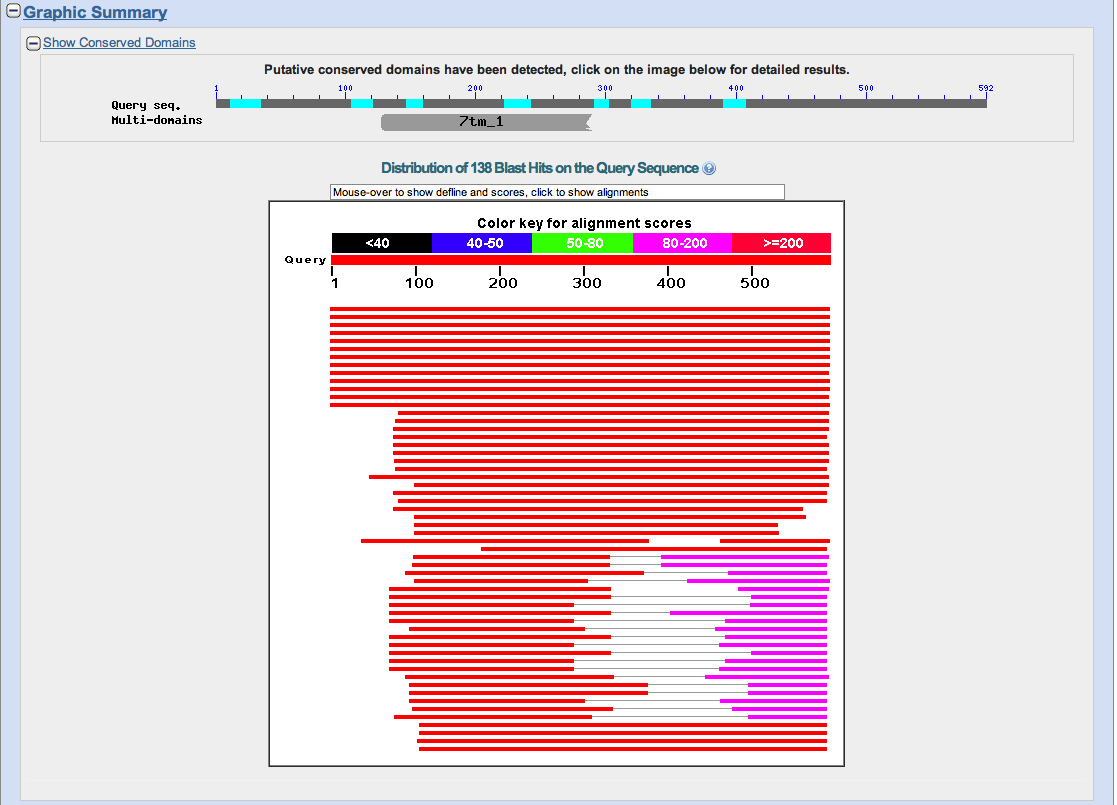
We use this peptide as the query sequence to search the nr protein database at NCBI using BLASTP. The results are shown below.

Notice at the top of the image that we have detected a conserved domain "7tm_1." Clicking on "7tm_1" gives us:
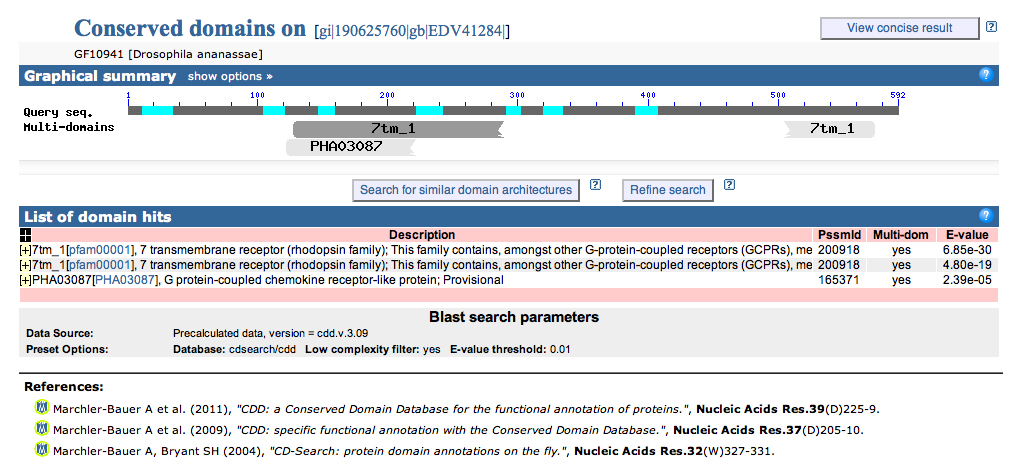
Clicking the first link gives us:
 |
7 transmembrane receptor (rhodopsin family)
This family contains, amongst other G-protein-coupled receptors (GCPRs), members of the opsin family, which have been considered to be typical members of the rhodopsin superfamily. They share several motifs, mainly the seven transmembrane helices, GCPRs of the rhodopsin superfamily. All opsins bind a chromophore, such as 11-cis-retinal. The function of most opsins other than the photoisomerases is split into two steps: light absorption and G-protein activation. Photoisomerases, on the other hand, are not coupled to G-proteins - they are thought to generate and supply the chromophore that is used by visual opsins. |
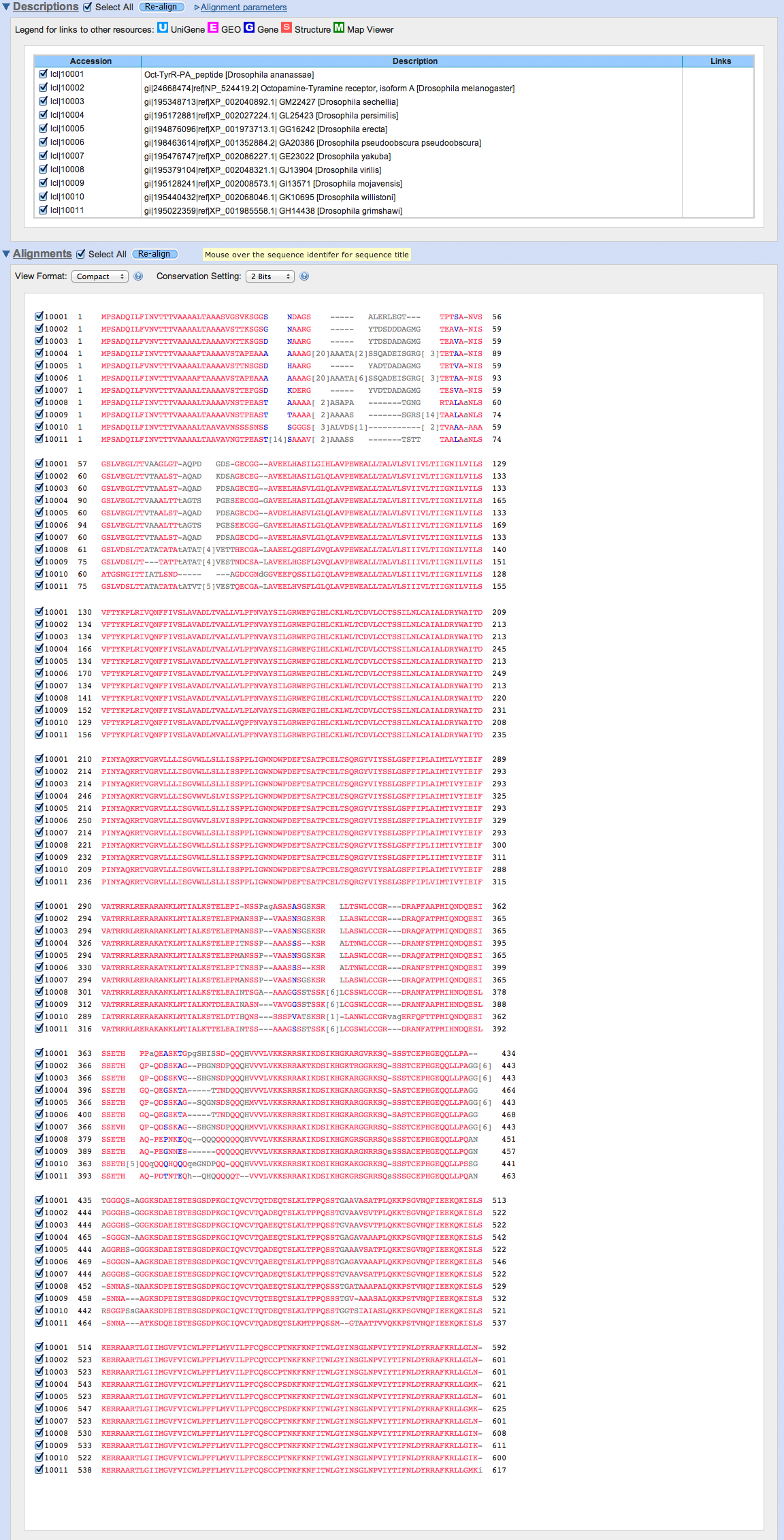
Our BLASTP output includes hits on very similar genes in related Drosophila species. We collect the sequences from the links in the alignments, gathering sequences in FASTA format to create the list below.
>Oct-TyrR-PA_peptide [Drosophila ananassae] MPSADQILFINVTTTVAAAALTAAASVGSVKSGGSNDAGSALERLEGTTPTSANVSGSLV EGLTTVAAGLGTAQPDGDSGECGGAVEELHASILGIHLAVPEWEALLTALVLSVIIVLTI IGNILVILSVFTYKPLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKL WLTCDVLCCTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPP LIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFIPLAIMTLVYIEIFVATRRRLRERA RANKLNTIALKSTELEPINSSPAGASASASGSKSRLLTSWLCCGRDRAPFAAPMIQNDQE SISSETHPPAQEASKTGPGSHISSDQQQHVVVLVKKSRRSKIKDSIKHGKARGVRKSQSS STCEPHGEQQLLPATGGGQSAGGKSDAEISTESGSDPKGCIQVCVTQTDEQTSLKLTPPQ SSTGAAVASATPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMY VILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN >gi|24668474|ref|NP_524419.2| Octopamine-Tyramine receptor, isoform A [Drosophila melanogaster] MPSADQILFVNVTTTVAAAALTAAAAVSTTKSGSGNAARGYTDSDDDAGMGTEAVANISGSLVEGLTTVT AALSTAQADKDSAGECEGAVEELHASILGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYKPL RIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWA ITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFI PLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDR AQFATPMIQNDQESISSETHQPQDSSKAGPHGNSDPQQQHVVVLVKKSRRAKTKDSIKHGKTRGGRKSQS SSTCEPHGEQQLLPAGGDGGSCQPGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQTSLKLTPPQS STGVAAVSVTPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILPFCQTCCP TNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN >gi|195348713|ref|XP_002040892.1| GM22427 [Drosophila sechellia] MPSADQILFVNVTTTVAAAALTAAAAVNTTKSGSDNAARGYTDSDADAGMGTEAVANISGSLVEGLTTVT AALSTAQADPDSAGECEGAVEELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYKPL RIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWA ITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFI PLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDR AQFATPMIQNDQESISSETHQPQDSSKVGSHGNSDPQQQHVVVLVKKSRRAKIKDSIKHGKARGGRKSQS SSTCEPHGEQQLLPAGGDGGRCQAGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQAEEQTSLKLTPPQS STGVAAVSVTPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILPFCQSCCP TNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN >gi|195172881|ref|XP_002027224.1| GL25423 [Drosophila persimilis] MPSADQILFINVTTTVAAAAFTAAAAVSTAPEAAAAAAAGGSTKSVTVVSAAAAAAAAAAAAATAASSSQ ADEISGRGASATETAANISGSLVEGLTTVAAALTTTAGTSPGESEECGGGAVEELHASILGLQLAVPEWE ALLTALVLSIIIVLTIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFG IHLCKLWLTCDVLCCTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWVLSLVISSPPLIGW NDWPDEFTSATPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERAKATKLNTIALKSTE LEPITNSSPAAASSSKSRALTNWLCCGRDRANFSTPMIQNDQESISSETHGQQEGSKTATTNDQQQHVVV LVKKSRRSKIKDSIKHGKARGGRKSQSASTCEPHGEQQLLPAGGSGGGNAAGKSDAEISTESGSDPKGCI QVCVTQAEEQTSLKLTPPQSSTGAGAVAAAPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVI CWLPFFLMYVILPFCQSCCPSDKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMK >gi|194876096|ref|XP_001973713.1| GG16242 [Drosophila erecta] MPSADQILFVNVTTTVAAAALTAAAAVSTTNSGSDHAARGYADTDADAGMGTETVANISGSLVEGLTTVT AALSTAQADPDSAGECDGAVDELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYKPL RIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWA ITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFI PLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDR AQFATPMIQNDQESISSETHQPQDSSKAGSQGNSDSQQQHMVVLVKKSRRAKIKDSIKHGKARGGRKSQS SSTCEPHGEQQLLPAGGDGGSCQAGGRHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQTSLKLTPPQS STGAAAVSATPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILPFCQSCCP TNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN >gi|198463614|ref|XP_001352884.2| GA20386 [Drosophila pseudoobscura pseudoobscura] MPSADQILFINVTTTVAAAAFTAAAAVSTAPEAAAAAAAGGSTKSVTVVSAAAAAAAAAAAAATATATGA SSSQADEISGRGASATETAANISGSLVEGLTTVAAALTTTAGTSPGESEECGGGAVEELHASILGLQLAV PEWEALLTALVLSIIIVLTIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGR WEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWVLSLVISSPP LIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERAKATKLNTIAL KSTELEPITNSSPAAASSSKSRALTNWLCCGRDRANFSTPMIQNDQESISSETHGQQEGSKTATTNDQQQ HVVVLVKKSRRSKIKDSIKHGKARGGRKSQSASTCEPHGEQQLLPAGGSGGGNAAGKSDAEISTESGSDP KGCIQVCVTQAEEQTSLKLTPPQSSTGAGAVAAAPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMG VFVICWLPFFLMYVILPFCQSCCPSDKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMK >gi|195476747|ref|XP_002086227.1| GE23022 [Drosophila yakuba] MPSADQILFVNVTTTVAAAALTAAAAVSTTEFGSDKDERGYVDTDADAGMGTESVANISGSLVEGLTTVT AALSTAQADPDSAGECDGAVEELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYKPL RIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWA ITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYSSLGSFFI PLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTELEPMANSSPVAASNSGSKSRLLASWLCCGRDR AQFATPMIQNDQESISSEVHQPQDSSKAGSHGNSDPQQQHMVVLVKKSRRAKIKDSIKHGKARGGRRSQS SSTCEPHGEQQLLPAGGDGVSCQAGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQTSLKLTPPQS STGVAAVSATPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILPFCQSCCP TNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN >gi|195379104|ref|XP_002048321.1| GJ13904 [Drosophila virilis] MPSADQILFINVTTTVAAAALTAAAAVNSTPEASTAAAAAKAASAPATGNGRTALAANLSGSLVDSLTTA TATATATATATAATIVETTHECGALAAEELQGSFLGVQLAVPEWEALLTALVLSIIIVLTIIGNILVILS VFTYKPLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAI ALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYS SLGSFFIPLIIMTIVYIEIFVATRRRLRERAKANKLNTIALKSTELEAINTSGAAAAGGSSTSSKARALG SLCSSWLCCGRDRANFATPMIHNDQESLSSETHAQPEPNKEQQQQQQQQQQQHVVVLVKKSRRAKIKDSI KHGKGRSGRRSQSSSSTCEPHGEQQLLPQANSNNASNAAKSDPEISTESGSDPKGCIQVCVTQAEEQTSL KLTPPQSSSTGATAAAPALQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILP FCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIN >gi|195128241|ref|XP_002008573.1| GI13571 [Drosophila mojavensis] MPSADQILFINVTTTVAAAALTAAAAVNSTPEASTTAAAATAAAAASSGRSVETAAATATVASTTTAALA ANLSGSLVDSLTTTATTTATATATAIVESTNDCSALAVEELHGSFLGVQLAVPEWEALLTALVLSIIIVL TIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLTVALLVLPLNVAYSILGRWEFGIHLCKLWLTCDVLC CTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSATPCE LTSQRGYVIYSSLGSFFIPLIIMTIVYIEIFVATRRRLRERAKANKLNTIALKNTDLEAINASNVAVGGS STSSKARALGSLCGSWLCCGRDRANFAAPMIHNDQESLSSETHAQPEGNNESQQQQQQHVVVLVKKSRRA KIKDSIKHGKARGNRRSQSSSSACEPHGEQQLLPQGNSNNAAGKSDPEISTESGSDPKGCIQVCVTQTEE QTSLKLTPPQSSSTGVAAASALQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYV ILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIK >gi|195440432|ref|XP_002068046.1| GK10695 [Drosophila willistoni] MPSADQILFINVTTTVAAAALTAAVAVNSSSSNSSSGGGSGNMALVDSLATTVAAAAAAATGSNGITTIA TLSNDAGDCGNDGGVEEFQSSILGIQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYKPLRIVQN FFIVSLAVADLTVALLVQPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSILNLCAIALDRYWAITDPI NYAQKRTVGRVLLLISGVWILSLLISSPPLIGWNDWPDEFTSATPCELTSQRGYVIYSALGSFFIPLAIM TIVYIEIFIATRRRLRERAKANKLNTIALKSTELDTIHQNSSSSPVATSKSRQLANWLCCGRVAGERFQF TTPMIQNDQESISSETHPHHHQQQQQQQHQQQQEGNDPQQQQQHVVVLVKKSRRSKIKDSIKHGKAKGGR KSQSSSTCEPHGEQQLLPSSGRSGGPSSGAAKSDPEISTESGSDPKGCIQVCITQTDEQTSLKLTPPQSS TGGTSIAIASLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPFFLMYVILPFCESCCPT NKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIK >gi|195022359|ref|XP_001985558.1| GH14438 [Drosophila grimshawi] MPSADQILFINVTTTVAAAALTAAVAVNGTPEASTGKASSPAAANGRSASAAAVAGAAASSTSTTTAALA ANLSGSLVDSLTTATATATATATVTTATAIVESTQECGALAVEELHVSFLGLQLAVPEWEALLTALVLSI IIVLTIIGNILVILSVFTYKPLRIVQNFFIVSLAVADLMVALLVLPFNVAYSILGRWEFGIHLCKLWLTC DVLCCTSSILNLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA TPCELTSQRGYVIYSSLGSFFIPLVIMTIVYIEIFVATRRRLRERAKANKLNTIALKTTELEAINTSSAA AGSSSTSSKARALGSLCGSWLCCGRDRANFATPMIHNDQESLSSETHAQPDTNTEQHQHQQQQQTVVVLV KKSRRAKIKDSIKHGKGRSGRRSQSSSSGCEPHGEQQLLPQANSNNAATKSDQEISTESGSDPKGCIQVC VTQADEQTSLKMTPPQSSMGTAATTVVQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVICWLPF FLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMKI
This list can be used as input to various multiple alignment programs. The first program that we will use is Clustal Omega. Here is the output.
CLUSTAL O(1.1.0) multiple sequence alignment
Oct-TyrR-PA_peptide MPSADQILFINVTTTVAAAALTAAASVGSVKSGGSNDAGSA-------------------
gi|24668474|ref|NP_524419.2| MPSADQILFVNVTTTVAAAALTAAAAVSTTKSGSGNAARGY-------------------
gi|195348713|ref|XP_002040892.1| MPSADQILFVNVTTTVAAAALTAAAAVNTTKSGSDNAARGY-------------------
gi|195172881|ref|XP_002027224.1| MPSADQILFINVTTTVAAAAFTAAAAVSTAPEAAAAAAAGGSTKSVTVVSAAAAAAAAAA
gi|194876096|ref|XP_001973713.1| MPSADQILFVNVTTTVAAAALTAAAAVSTTNSGSDHAARGY-------------------
gi|198463614|ref|XP_001352884.2| MPSADQILFINVTTTVAAAAFTAAAAVSTAPEAAAAAAAGGSTKSVTVVSAAAAAAAAAA
gi|195476747|ref|XP_002086227.1| MPSADQILFVNVTTTVAAAALTAAAAVSTTEFGSDKDERGY-------------------
gi|195379104|ref|XP_002048321.1| MPSADQILFINVTTTVAAAALTAAAAVNSTPEASTAAAAAKAASAPATG-----------
gi|195128241|ref|XP_002008573.1| MPSADQILFINVTTTVAAAALTAAAAVNSTPEASTTAAAATAAAAASSGRSVET--AAA-
gi|195440432|ref|XP_002068046.1| MPSADQILFINVTTTVAAAALTAAVAVNSSSSNSSSGGGSGNM---ALVDSLATTVAAAA
gi|195022359|ref|XP_001985558.1| MPSADQILFINVTTTVAAAALTAAVAVNGTPEASTGKASSPAAANGRSASAAAV--AGA-
*********:**********:***.:* . .
Oct-TyrR-PA_peptide ---------------LER---LEGTTPTSANVSGSLVEGLTTVAAGLGTA-----QP-DG
gi|24668474|ref|NP_524419.2| ---------------TDSDDDAGMGTEAVANISGSLVEGLTTVTAALSTA-----QA-DK
gi|195348713|ref|XP_002040892.1| ---------------TDSDADAGMGTEAVANISGSLVEGLTTVTAALSTA-----QA-DP
gi|195172881|ref|XP_002027224.1| ----AAATAASSSQADEISGRGASATETAANISGSLVEGLTTVAAALTTT-----AGTSP
gi|194876096|ref|XP_001973713.1| ---------------ADTDADAGMGTETVANISGSLVEGLTTVTAALSTA-----QA-DP
gi|198463614|ref|XP_001352884.2| AAATATATGASSSQADEISGRGASATETAANISGSLVEGLTTVAAALTTT-----AGTSP
gi|195476747|ref|XP_002086227.1| ---------------VDTDADAGMGTESVANISGSLVEGLTTVTAALSTA-----QA-DP
gi|195379104|ref|XP_002048321.1| ----------------------NGRTALAANLSGSLVDSLTTATATATATATA-TAATIV
gi|195128241|ref|XP_002008573.1| ----------------TATVASTTTAALAANLSGSLVDSLTTT---ATTTATA-TATAIV
gi|195440432|ref|XP_002068046.1| AA-------------------------------ATGSNGITTI-------------ATLS
gi|195022359|ref|XP_001985558.1| ----------------AASSTSTTTAALAANLSGSLVDSLTTATATATATATVTTATAIV
.: :.:**
Oct-TyrR-PA_peptide D-SGEC--GGAVEELHASILGIHLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYK
gi|24668474|ref|NP_524419.2| DSAGEC--EGAVEELHASILGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYK
gi|195348713|ref|XP_002040892.1| DSAGEC--EGAVEELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYK
gi|195172881|ref|XP_002027224.1| GESEECG-GGAVEELHASILGLQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
gi|194876096|ref|XP_001973713.1| DSAGEC--DGAVDELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYK
gi|198463614|ref|XP_001352884.2| GESEECG-GGAVEELHASILGLQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
gi|195476747|ref|XP_002086227.1| DSAGEC--DGAVEELHASVLGLQLAVPEWEALLTALVLSVIIVLTIIGNILVILSVFTYK
gi|195379104|ref|XP_002048321.1| ETTHECG-ALAAEELQGSFLGVQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
gi|195128241|ref|XP_002008573.1| ESTNDCS-ALAVEELHGSFLGVQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
gi|195440432|ref|XP_002068046.1| NDAGDCGNDGGVEEFQSSILGIQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
gi|195022359|ref|XP_001985558.1| ESTQECG-ALAVEELHVSFLGLQLAVPEWEALLTALVLSIIIVLTIIGNILVILSVFTYK
: :* ..:*:: *.**::****************:********************
Oct-TyrR-PA_peptide PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|24668474|ref|NP_524419.2| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195348713|ref|XP_002040892.1| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195172881|ref|XP_002027224.1| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|194876096|ref|XP_001973713.1| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|198463614|ref|XP_001352884.2| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195476747|ref|XP_002086227.1| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195379104|ref|XP_002048321.1| PLRIVQNFFIVSLAVADLTVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195128241|ref|XP_002008573.1| PLRIVQNFFIVSLAVADLTVALLVLPLNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195440432|ref|XP_002068046.1| PLRIVQNFFIVSLAVADLTVALLVQPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
gi|195022359|ref|XP_001985558.1| PLRIVQNFFIVSLAVADLMVALLVLPFNVAYSILGRWEFGIHLCKLWLTCDVLCCTSSIL
****************** ***** *:*********************************
Oct-TyrR-PA_peptide NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|24668474|ref|NP_524419.2| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|195348713|ref|XP_002040892.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|195172881|ref|XP_002027224.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWVLSLVISSPPLIGWNDWPDEFTSA
gi|194876096|ref|XP_001973713.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|198463614|ref|XP_001352884.2| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWVLSLVISSPPLIGWNDWPDEFTSA
gi|195476747|ref|XP_002086227.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|195379104|ref|XP_002048321.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|195128241|ref|XP_002008573.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
gi|195440432|ref|XP_002068046.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWILSLLISSPPLIGWNDWPDEFTSA
gi|195022359|ref|XP_001985558.1| NLCAIALDRYWAITDPINYAQKRTVGRVLLLISGVWLLSLLISSPPLIGWNDWPDEFTSA
************************************:***:*******************
Oct-TyrR-PA_peptide TPCELTSQRGYVIYSSLGSFFIPLAIMTLVYIEIFVATRRRLRERARANKLNTIALKSTE
gi|24668474|ref|NP_524419.2| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTE
gi|195348713|ref|XP_002040892.1| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTE
gi|195172881|ref|XP_002027224.1| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERAKATKLNTIALKSTE
gi|194876096|ref|XP_001973713.1| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTE
gi|198463614|ref|XP_001352884.2| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERAKATKLNTIALKSTE
gi|195476747|ref|XP_002086227.1| TPCELTSQRGYVIYSSLGSFFIPLAIMTIVYIEIFVATRRRLRERARANKLNTIALKSTE
gi|195379104|ref|XP_002048321.1| TPCELTSQRGYVIYSSLGSFFIPLIIMTIVYIEIFVATRRRLRERAKANKLNTIALKSTE
gi|195128241|ref|XP_002008573.1| TPCELTSQRGYVIYSSLGSFFIPLIIMTIVYIEIFVATRRRLRERAKANKLNTIALKNTD
gi|195440432|ref|XP_002068046.1| TPCELTSQRGYVIYSALGSFFIPLAIMTIVYIEIFIATRRRLRERAKANKLNTIALKSTE
gi|195022359|ref|XP_001985558.1| TPCELTSQRGYVIYSSLGSFFIPLVIMTIVYIEIFVATRRRLRERAKANKLNTIALKTTE
***************:******** ***:******:**********:*.********.*:
Oct-TyrR-PA_peptide LEPINSSPAGASASA-SGSK----SRLLTSWLCCGR---DRAPFAAPMIQNDQESISSET
gi|24668474|ref|NP_524419.2| LEPMANSSP-VAASN-SGSK----SRLLASWLCCGR---DRAQFATPMIQNDQESISSET
gi|195348713|ref|XP_002040892.1| LEPMANSSP-VAASN-SGSK----SRLLASWLCCGR---DRAQFATPMIQNDQESISSET
gi|195172881|ref|XP_002027224.1| LEPITNSSPAA---A-SSSK----SRALTNWLCCGR---DRANFSTPMIQNDQESISSET
gi|194876096|ref|XP_001973713.1| LEPMANSSP-VAASN-SGSK----SRLLASWLCCGR---DRAQFATPMIQNDQESISSET
gi|198463614|ref|XP_001352884.2| LEPITNSSPAA---A-SSSK----SRALTNWLCCGR---DRANFSTPMIQNDQESISSET
gi|195476747|ref|XP_002086227.1| LEPMANSSP-VAASN-SGSK----SRLLASWLCCGR---DRAQFATPMIQNDQESISSEV
gi|195379104|ref|XP_002048321.1| LEAINTSGAAAAGGSSTSSKARALGSLCSSWLCCGR---DRANFATPMIHNDQESLSSET
gi|195128241|ref|XP_002008573.1| LEAINASNV-AVGGSSTSSKARALGSLCGSWLCCGR---DRANFAAPMIHNDQESLSSET
gi|195440432|ref|XP_002068046.1| LDTIHQNSSSS--PV-ATSK----SRQLANWLCCGRVAGERFQFTTPMIQNDQESISSET
gi|195022359|ref|XP_001985558.1| LEAINTSSA-AAGSSSTSSKARALGSLCGSWLCCGR---DRANFATPMIHNDQESLSSET
*: : . : ** . .****** :* *::***:*****:***.
Oct-TyrR-PA_peptide HPPAQEASK-----TGPGSHISSDQQQHVVVLVKKSRRSKIKDSIKHGKARGVRKSQS-S
gi|24668474|ref|NP_524419.2| HQPQDS-SK-----AGPHG-NSDPQQQHVVVLVKKSRRAKTKDSIKHGKTRGGRKSQS-S
gi|195348713|ref|XP_002040892.1| HQPQDS-SK-----VGSHG-NSDPQQQHVVVLVKKSRRAKIKDSIKHGKARGGRKSQS-S
gi|195172881|ref|XP_002027224.1| HGQQEG----------SKTATTNDQQQHVVVLVKKSRRSKIKDSIKHGKARGGRKSQS-A
gi|194876096|ref|XP_001973713.1| HQPQDS-SK-----AGSQG-NSDSQQQHMVVLVKKSRRAKIKDSIKHGKARGGRKSQS-S
gi|198463614|ref|XP_001352884.2| HGQQEG----------SKTATTNDQQQHVVVLVKKSRRSKIKDSIKHGKARGGRKSQS-A
gi|195476747|ref|XP_002086227.1| HQPQDS-SK-----AGSHG-NSDPQQQHMVVLVKKSRRAKIKDSIKHGKARGGRRSQS-S
gi|195379104|ref|XP_002048321.1| HAQPEPNKEQQQ-------QQQQQQQQHVVVLVKKSRRAKIKDSIKHGKGRSGRRSQSSS
gi|195128241|ref|XP_002008573.1| HAQPEGNNESQ-----------QQQQQHVVVLVKKSRRAKIKDSIKHGKARGNRRSQSSS
gi|195440432|ref|XP_002068046.1| HPHHHQQQQQQQHQQQQEGNDPQQQQQHVVVLVKKSRRSKIKDSIKHGKAKGGRKSQS-S
gi|195022359|ref|XP_001985558.1| HAQPDTNTEQHQ-------H--QQQQQTVVVLVKKSRRAKIKDSIKHGKGRSGRRSQSSS
* . . *** :*********:* ******** :. *:*** :
Oct-TyrR-PA_peptide STCEPHGEQQLLPATG--------GGQSAGGKSDAEISTESGSDPKGCIQVCVTQTDEQT
gi|24668474|ref|NP_524419.2| STCEPHGEQQLLPAGGDGGSCQPGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQT
gi|195348713|ref|XP_002040892.1| STCEPHGEQQLLPAGGDGGRCQAGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQAEEQT
gi|195172881|ref|XP_002027224.1| STCEPHGEQQLLPAGGSGG-------GNAAGKSDAEISTESGSDPKGCIQVCVTQAEEQT
gi|194876096|ref|XP_001973713.1| STCEPHGEQQLLPAGGDGGSCQAGGRHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQT
gi|198463614|ref|XP_001352884.2| STCEPHGEQQLLPAGGSGG-------GNAAGKSDAEISTESGSDPKGCIQVCVTQAEEQT
gi|195476747|ref|XP_002086227.1| STCEPHGEQQLLPAGGDGVSCQAGGGHSGGGKSDAEISTESGSDPKGCIQVCVTQADEQT
gi|195379104|ref|XP_002048321.1| STCEPHGEQQLLPQANSNN-------ASNAAKSDPEISTESGSDPKGCIQVCVTQAEEQT
gi|195128241|ref|XP_002008573.1| SACEPHGEQQLLPQGNSNN-------A--AGKSDPEISTESGSDPKGCIQVCVTQTEEQT
gi|195440432|ref|XP_002068046.1| STCEPHGEQQLLPSSGRSG-----GPSSGAAKSDPEISTESGSDPKGCIQVCITQTDEQT
gi|195022359|ref|XP_001985558.1| SGCEPHGEQQLLPQANSNN-------A--ATKSDQEISTESGSDPKGCIQVCVTQADEQT
* *********** . *** *****************:**::***
Oct-TyrR-PA_peptide SLKLTPPQSSTGAAVASATPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|24668474|ref|NP_524419.2| SLKLTPPQSSTGVAAVSVTPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195348713|ref|XP_002040892.1| SLKLTPPQSSTGVAAVSVTPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195172881|ref|XP_002027224.1| SLKLTPPQSSTGAGAVAAAPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|194876096|ref|XP_001973713.1| SLKLTPPQSSTGAAAVSATPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|198463614|ref|XP_001352884.2| SLKLTPPQSSTGAGAVAAAPLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195476747|ref|XP_002086227.1| SLKLTPPQSSTGVAAVSATPLQKKTSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195379104|ref|XP_002048321.1| SLKLTPPQSSSTGATAAAPALQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195128241|ref|XP_002008573.1| SLKLTPPQSSSTGVA-AASALQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195440432|ref|XP_002068046.1| SLKLTPPQSSTGGTSIAIASLQKKPSGVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
gi|195022359|ref|XP_001985558.1| SLKMTPPQSSMGT--AATTVVQKKPSTVNQFIEEKQKISLSKERRAARTLGIIMGVFVIC
***:****** : :*** * *********************************
Oct-TyrR-PA_peptide WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
gi|24668474|ref|NP_524419.2| WLPFFLMYVILPFCQTCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
gi|195348713|ref|XP_002040892.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
gi|195172881|ref|XP_002027224.1| WLPFFLMYVILPFCQSCCPSDKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMK
gi|194876096|ref|XP_001973713.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
gi|198463614|ref|XP_001352884.2| WLPFFLMYVILPFCQSCCPSDKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMK
gi|195476747|ref|XP_002086227.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGLN
gi|195379104|ref|XP_002048321.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIN
gi|195128241|ref|XP_002008573.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIK
gi|195440432|ref|XP_002068046.1| WLPFFLMYVILPFCESCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGIK
gi|195022359|ref|XP_001985558.1| WLPFFLMYVILPFCQSCCPTNKFKNFITWLGYINSGLNPVIYTIFNLDYRRAFKRLLGMK
**************::***::*************************************::
Oct-TyrR-PA_peptide -
gi|24668474|ref|NP_524419.2| -
gi|195348713|ref|XP_002040892.1| -
gi|195172881|ref|XP_002027224.1| -
gi|194876096|ref|XP_001973713.1| -
gi|198463614|ref|XP_001352884.2| -
gi|195476747|ref|XP_002086227.1| -
gi|195379104|ref|XP_002048321.1| -
gi|195128241|ref|XP_002008573.1| -
gi|195440432|ref|XP_002068046.1| -
gi|195022359|ref|XP_001985558.1| I
We also use the COBALT program at NCBI. The output is shown below.
We also use the MUSCLE program. The output is shown below.

The proteins that we have used are all closely related. All three programs have given us similar results. The portion of the protein identified as a conserved transmembrane domain common to G-protein-coupled receptors (around 120-300 and 500-575) are highly conserved in the group of proteins that we have analyzed. Notice that the sequences near the amino terminus are highly divergent.
You may find these programs useful in the analysis of your gene models.