Summary:
D. ananassae fosmid 2602K20 encodes an ortholog of D. melanogaster CG7140. There is an additional gene match with D. melanogaster CG43392.
BLASTX analysis:
Using the fosmid_2602K20, a 41165 letter long fosmid, from from the src folder in the GEP project file for D. ananassae as the query sequence, I went to the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and performed a BLASTX search of Non-redundant protein sequences restricted to D. melanogaster.

These results suggest that there are two possible gene matches on the fosmid (2602k20) with protein sequence similarity to the D. melanogaster protein set.
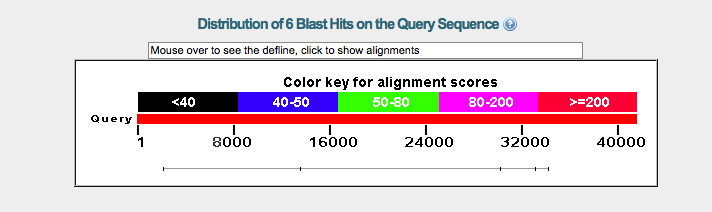
The graph in figure 1 depicts a small potential gene region in green. The alignment score for this gene is somewhat weak. There is a much stronger alignment score for the gene on the right side of the graph, this gene has additional isoforms that also show high alignment scores with the fosmid_2602K20.
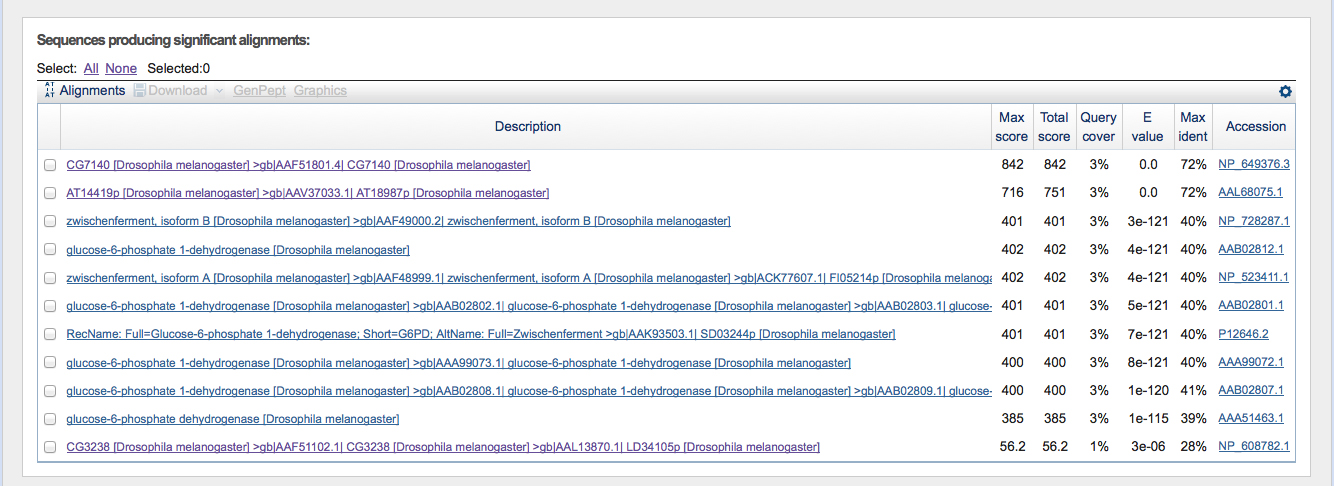
Using the E values given in the table in figure 2, I investigated the potential genes using FlyBase.
FlyBase revealed that CG7140 is located on chromosome 3L. This is a good potential match, as we know that the fosmid came from the 3L chromosome in D. ananassae. Using FlyBase to check AT14419p shows that it is an isoform of CG7140.
The zwischenferment gene (Zw) is located on the X chromosome and therefore it can be excluded from this analysis as a potential gene match. All of the remaining glucose-6-phosphate genes are isoforms of the Zw gene and can be excluded as potential matches as well.
CG3238 has a lower, yet still mildly significant alignment score. FlyBase confirmed that this gene is located on chromosome 2L. Therefore this gene can be excluded.
Alignment:
NP_649376.3:
Using the frame +3 we find that CG7140 (NP_649376.3) starts at 1 and is continuous to 522. The query, fosmid_2602k20, starts at 34557 and is continuous to 36122. The expect value is 0.0, the identity is 72%, and the match is 86% positive.
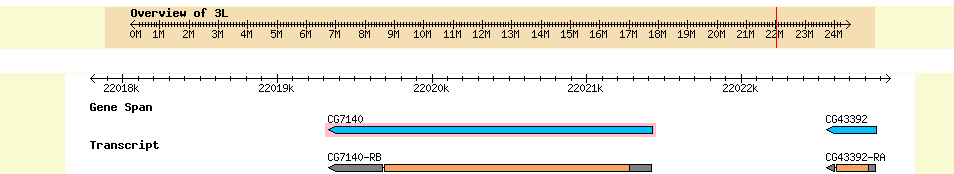
In summary, the evidence thus far shows that there is a gene on our D. ananassae fosmid_2602K20, that is an ortholog of the CG7140 gene found in D. melanogaster. This gene is a protein coding gene predicted to function in glucose-6-dehydrogenase activity and take part in glucose metabolism. A screen shot from FlyBase GBrowse is shown below in figure 3.
Next I used the Gene Record Finder at GEP to acquire the protein sequences from CG7140.
I then performed a BLASTX comparing the above sequence to the query fosmid_2602K20. The graph below (figure 4) shows that a small part of the query fosmid_2602K20 matched very well.
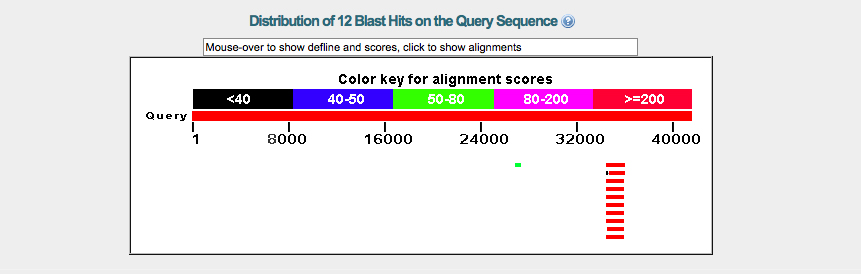
This area on the +3 reading frame spans from 34557 to 36122. The expect value is zero and all other values confirm that this is a significant match between the D. melanogaster gene CG7140 and a gene on our query fosmid.
BLAST analysis by GEP:
Using the GEP UCSC Genome browser for the D. ananassae genome, with the assembly set to the Jan. 2013 (GEP/3L Reference) for fosmid_2602K20. I was able to identify an additional potential gene. By adjusting the track settings to filter the score range to a minimum of zero and a max of 1000 I was able to see another potential gene match!
Figure 5
CG43392 is cross-referenced through FlyBase ID: (FBpp0300405) as a polypeptide sequence in D. melanogaster. This polypeptide sequence is located on chromosome 3L.
Alignment:
Using the +2 frame we find that CG43392-PA starts at 1 and is continuous through 74 in D. melanogaster. The query sequence lies between 33170 and 33385. The expect value is significant at 6.0e-13! The identity match is only 41%, but by referring to the modENCODE RNA-Seq section of figure 5 we can see that the sequence is transcribed.
Next I used the gene record finder to obtain the protein sequences for CG43392 and compared them to the query fosmid using BLASTX. See figure 6 below.
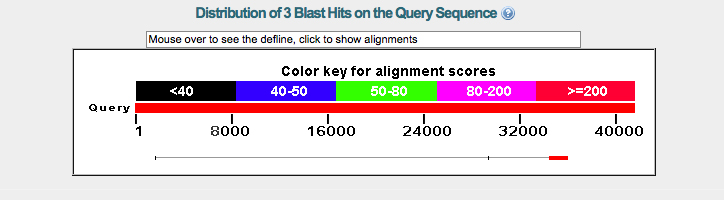
According to BLASTX there is no significant relationship between the query fosmid and CG43392.
I returned to GEP UCSC Genome browser to zoom in on CG43392. By setting the base position to "full" I was able to identify a start codon at 33,170 (ATG) and the gene runs until 33,385, just before a stop codon which starts at 33,404 (TAA) on the +2 reading frame. The gene appears to be an ortholog to CG43392 which transcribes an unknown polypeptide sequence in D. melanogaster.
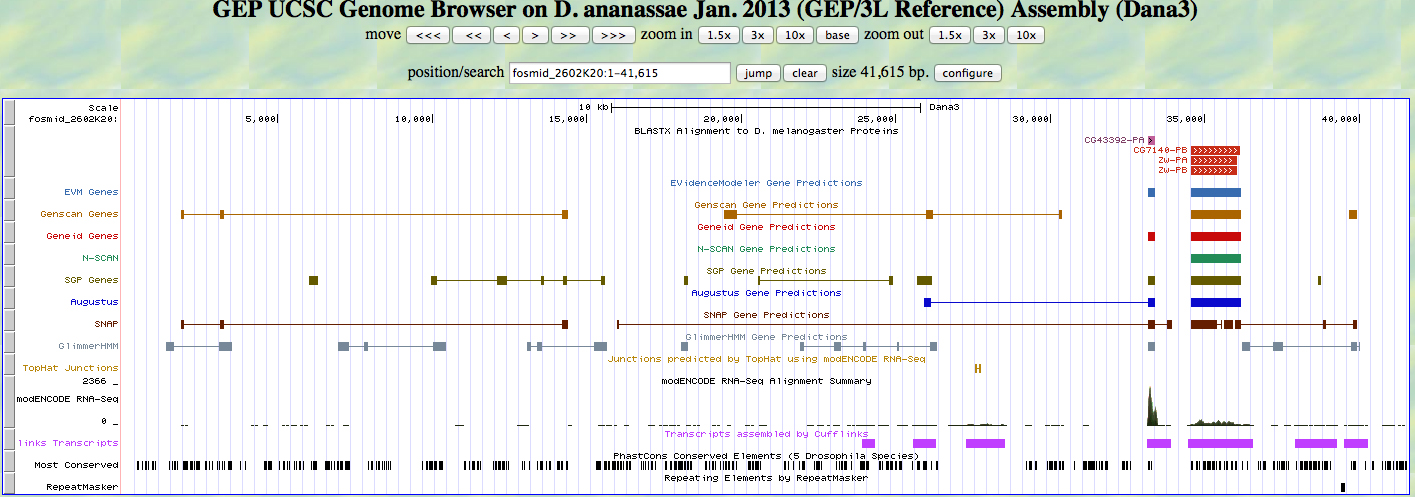
Next I used GEP UCSC Genome browser to closely examine the characteristics of CG7140. With the base position setting on "full" I was able to identify a start codon at 34,557 (ATG) and the gene runs until 36,146. There is a stop codon located at 36,164 (TAG) on the +3 reading frame (as predicted from the NCBI BLASTX analysis).