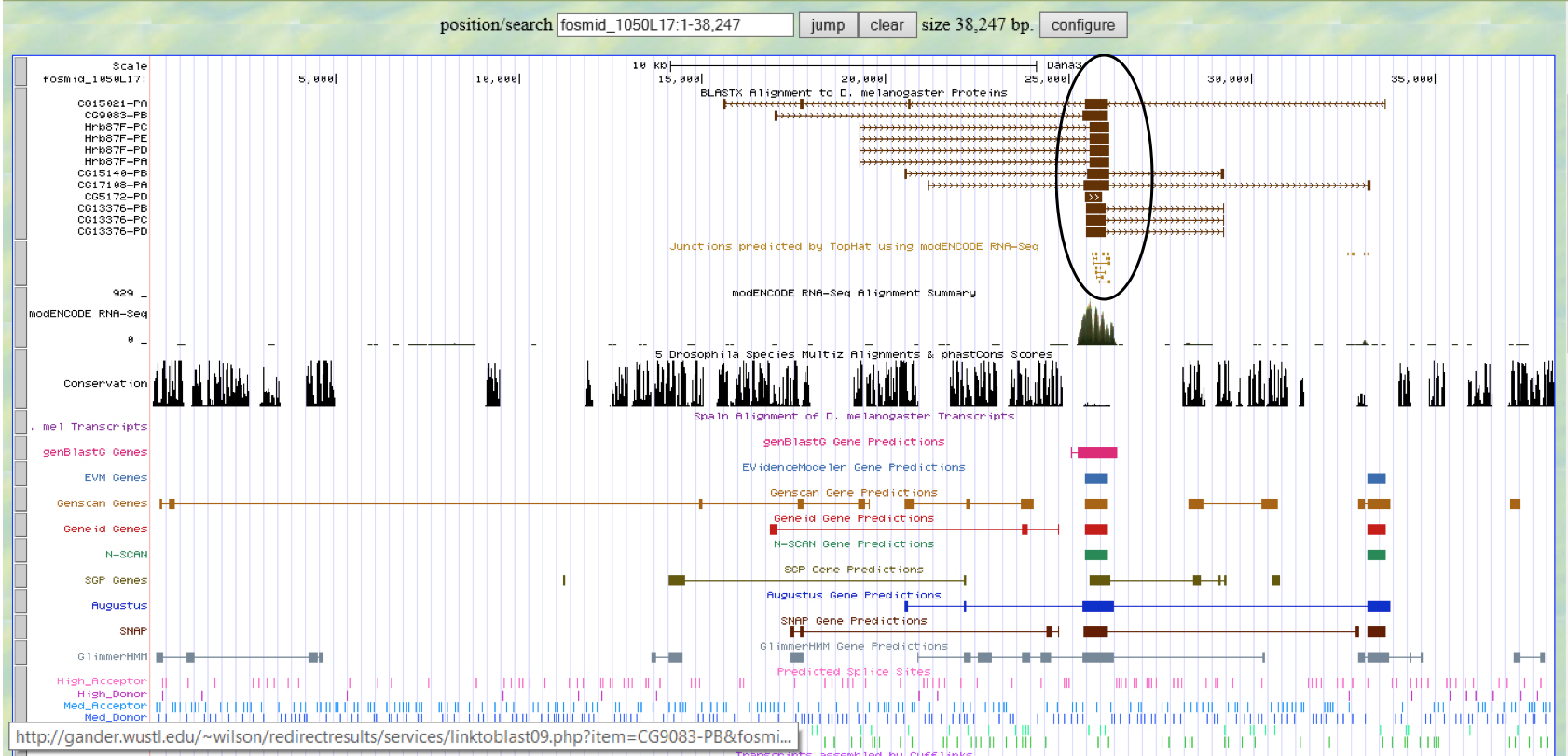
Summary: According to blastx analysis considering E-values, only two genes are possibly significant: TPA: HDC10038 (accession ID: DAA02974.1) and CG2982 (accession ID: NP_572160.1). However, according to GENSCAN analysis, only predicted peptide 4 has a low value of 3 e-5. However 3e-5 is still a rather high value. One thing we know with certainty is that CG2982 (accession ID: NP_572160.1) cannot be one of the genes in the fosmid due to the fact that GENSCAN rarely does not leave out genes, it only includes extra ones. GENSCAN did not show the presence of CG2982. UCSC Genome Browser analysis shows mRNA transcripts from actual experiments around the region of 26k to 27k kb which is near the region of the two later segments of the gene TPA: HDC10038 (accession ID: DAA02974.1) ~ 28k kb. However, based on mediocre relationships, we can only conclude the gene on the fosmid 1050L17 is possibly TPA: HDC10038 (accession ID: DAA02974.1) has which two sequences.
D. ananassae fosmid 1050L17 (using the file name fosmid_1050L17.fasta):

which gives:
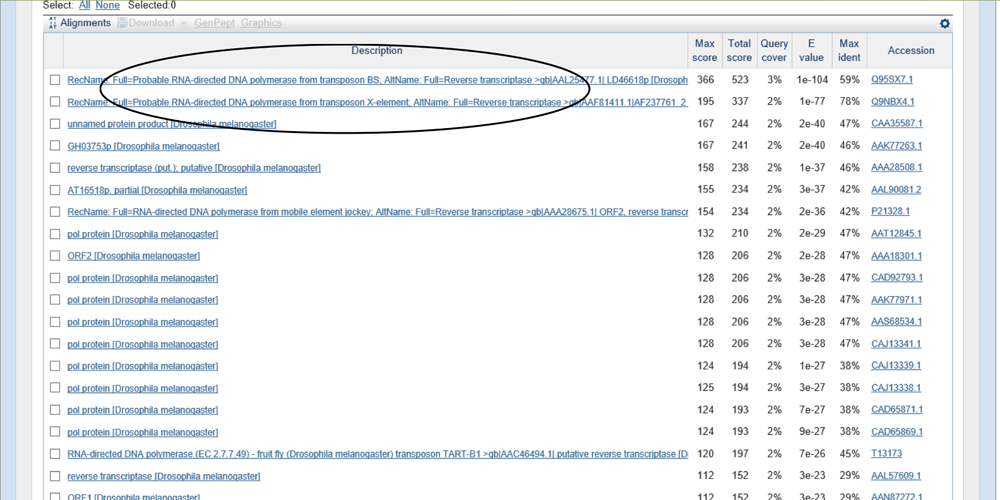
The alignments show results of RNA -directed DNA polymerase which calls for the use of the file fosmid_1050L17.fasta.masked using the same parameters:
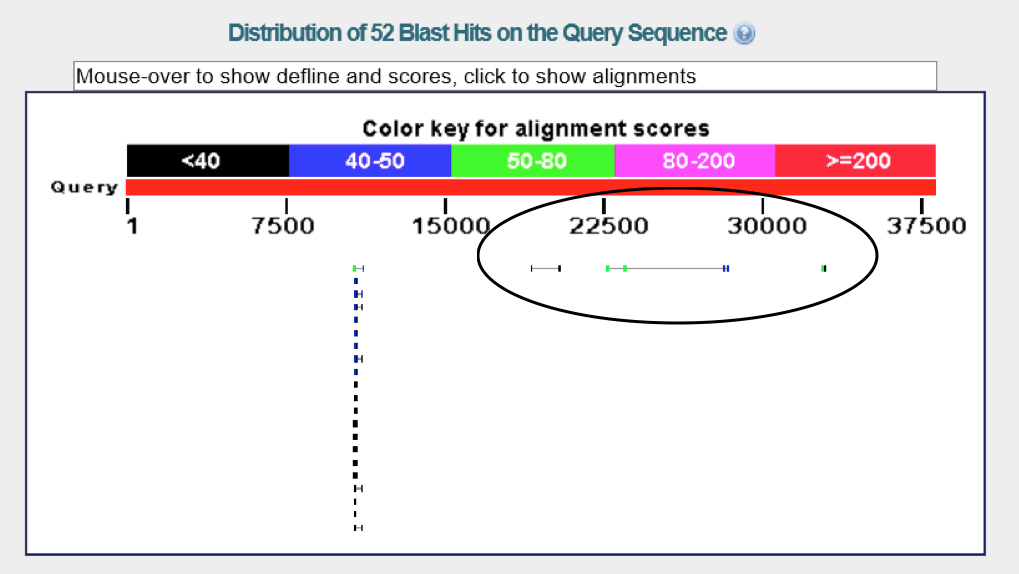
There are four genes on the fosmid. However, the gene at ~10k kb is the probable RNA directed DNA polymerase. Therefore, we will be looking at the genes from 17k to 33k and analyzing its validity from hereon.
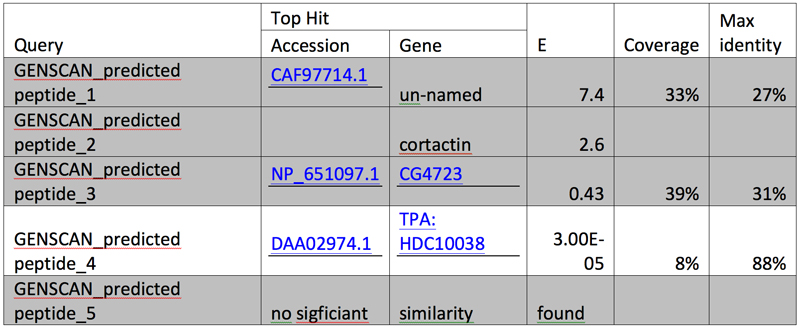
After the three genes are tabulated in an increasing order of their respective segments and alignments with E values larger than e-10 are grayed out, we have:
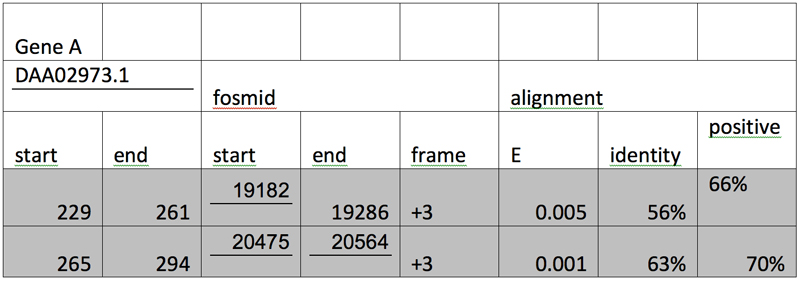
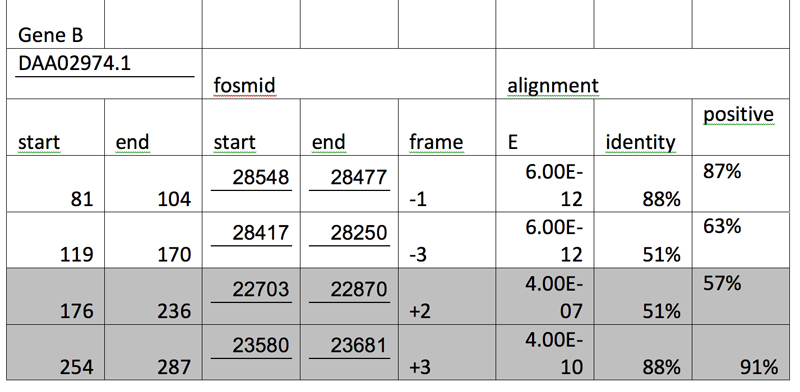
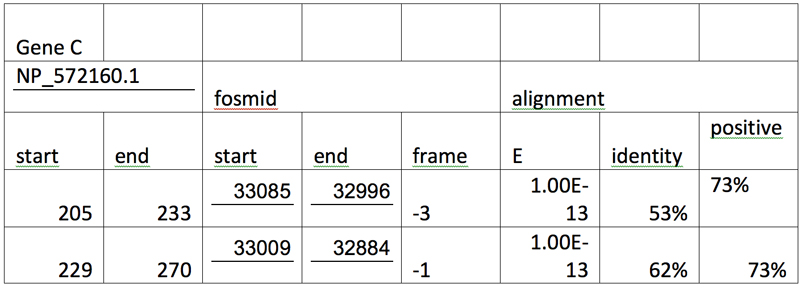
Genscan Analysis:
>fosmid_1050L17|GENSCAN_predicted_peptide_1|246_aa MVLAFPSTLPASTLPSAWVLRDRSPCSARYTDTDKVIYTQAGTRSHAHARTATVVIFVAV LQAMGSGRRGAGAAGARQRKAGLLWVSLRRFLGKSEHDQWLFVRDRGDRQCYGNKARPTT LPGVAVKNQRTDKLQNMVQEPKVGRLGEYGCGCECGFGFGQGHGCGMVQCIQFRSAHIQG PGLRFRSGIWPTVYLYLNLDCFANCQHPQKRQERPGRRQGGGGRSENWKRVCGPAATVNF HGMRFA >fosmid_1050L17|GENSCAN_predicted_peptide_2|231_aa MDCGRRSARLSGFPMTTMRDVCQQRHPDEALGWRSGVRGWDLGLGDRGLGVTGGKWAKEA DTGSASSMKENYEALFIVYAHYSDPSVNMRNWGSGGKTLPEKCKSVEGNRGGLWRTRIGD KFDLHTPQDHIHCEVQVQVQAQAHGQERMRDNNAECCASQLPRTHKQLCAGATRSPASRG NRNTIEAVWKSEVEHPCKRLSVTWSEGLVEHSPTKSGKHPSSATATPRGSH >fosmid_1050L17|GENSCAN_predicted_peptide_3|212_aa MEALEALEDLEDQVAQEDLVGLEDPEDLEGMEDMEGMEVTEGMEDLEELEDLPLLEMAVV EMLAAENLVVEMPAVENLVVEMPAVENPVVEMSAMENLVVEMTEVEMSVMQNLVEEMPAV EMPVVEMPAVENLVVEMSVVEIPVVENLVVEMPAVEMPVVEMQVVEMSALENLVVEMAVV EMPAVENLVVEIPAVENLVVEMPAVENLALEV >fosmid_1050L17|GENSCAN_predicted_peptide_4|275_aa MIQDSPEEKCMSIKKRKPNCWHTIWKSNPASPEACQPSPIHPVSIKVLGTNDSVSGFNLL ARIFGIFIVICFDLCMGVVVWALMKRKKPKGSHEDANALTRGNTCLSASGSRFQGSPRAS HDTFPASVGHGGSKNQILPVPPFLNAASRVSPIAAPPPFQPPSSFWLRRHRHPRGRSPAY CVANDRKFKRMPLCSSLAAPEAAEGKNICGSNKNTHNNKVLCFIQFCSLSWLGAGLVGFA LVSCVYPAAIISVLNLISSLTHQRQRQVKRSLPPT >fosmid_1050L17|GENSCAN_predicted_peptide_5|278_aa MVVNSSWWRISISSIILNEISSAKYLGVWRYIWQAFFSQKLPKNSAGRLDGIEDPLEDAA SFLNAVHLMSAAEVVGPLAAASFNLSLLDLALDLEVPGLGLIFSAAGDFEPLAGRPAGLG KEMQLVISMSMAGETPLALGPGLFLASSLRRGLRAFDFADPPALDSGAALAASVGFGDLA LASSGQEATILRFNGQWSAPVSPVTGSVCTRCTPGHGDAALDARITRSWARPFGGILGIG WRCPVVVGISGKGLLLLAANTDRPVVVVEVAFFAYFYS
According to GENSCAN, the gene TPA: HDC10038 is the closest significant gene having an E value of 3 e-5.
UCSC Genome Browser analysis: