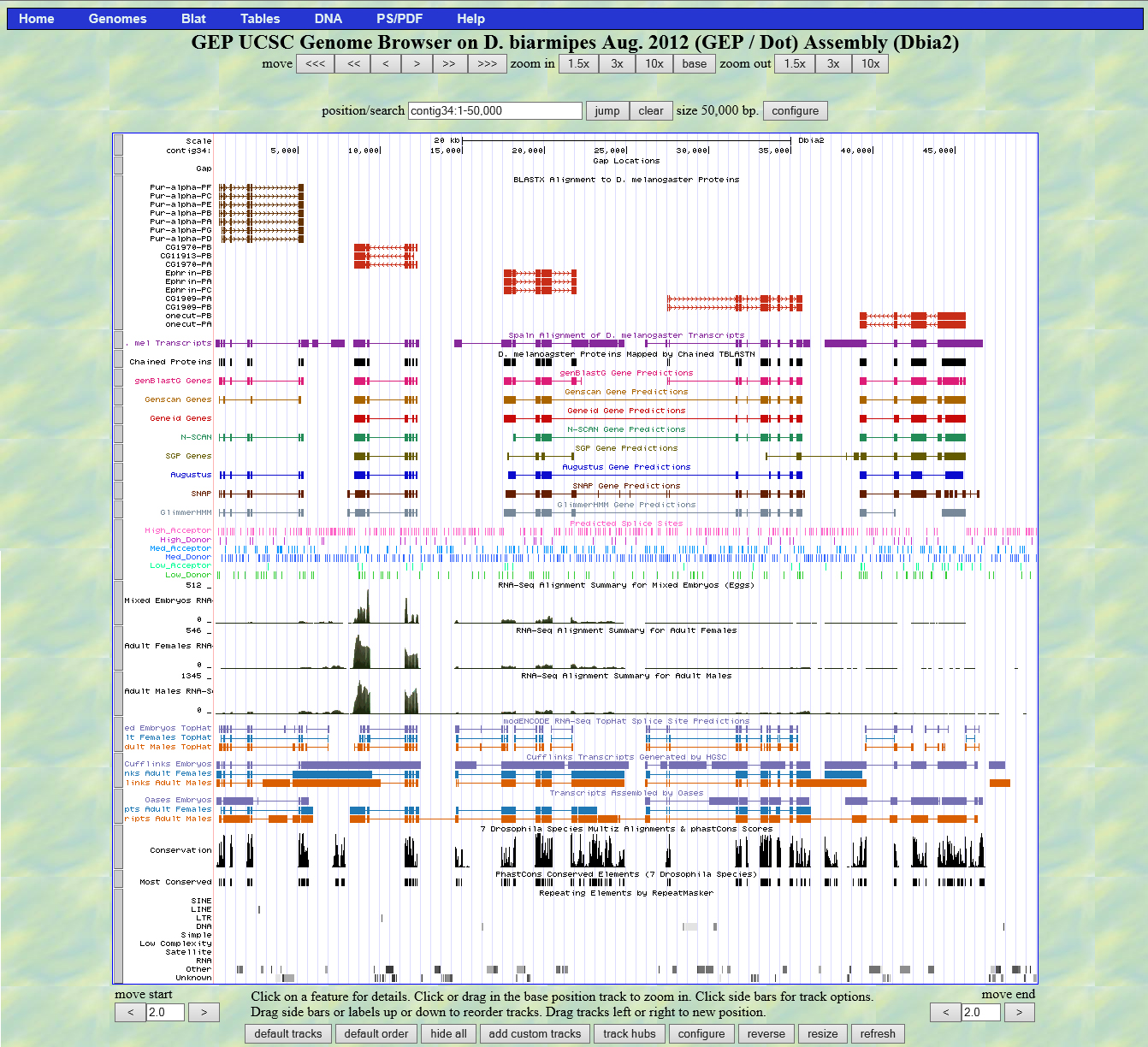
Summary:
D. biarmipes fosmid contig 34 appears to encode five genes. These genes are orthologs of D. melanogaster genes Pur-alpha, CG1970, Ephrin, CG1909, and Onecut.
BLASTX analysis:
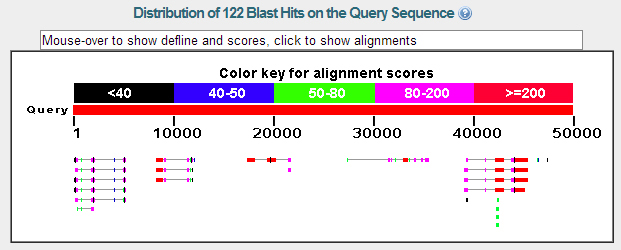
A BLASTX analysis using the NCBI database and the contig 34 sequence as a query revealed five good matches to proteins in the D. melanogaster set. There were good matches on several isoforms of the genes so I have combined the isoforms into single the single genes listed above in the summary.

A BLASTX conducted through FlyBase using the same contig 34 sequence resulted in similar data indicating the presence of the five genes with good matches to the same five genes previously listed.

Alignments:
I tabulated alignments on the gene matches obtained through the BLASTX analysis attempting to focus on the most significant scores and sorted the tables according to the order of the protein sequence. Tables are matched to the ortholog gene identified. Frame polarity identifies the reading frame as well as the strand the gene is located on with negative values occurring on the minus strand and reading in the reverse direction on the fosmid.
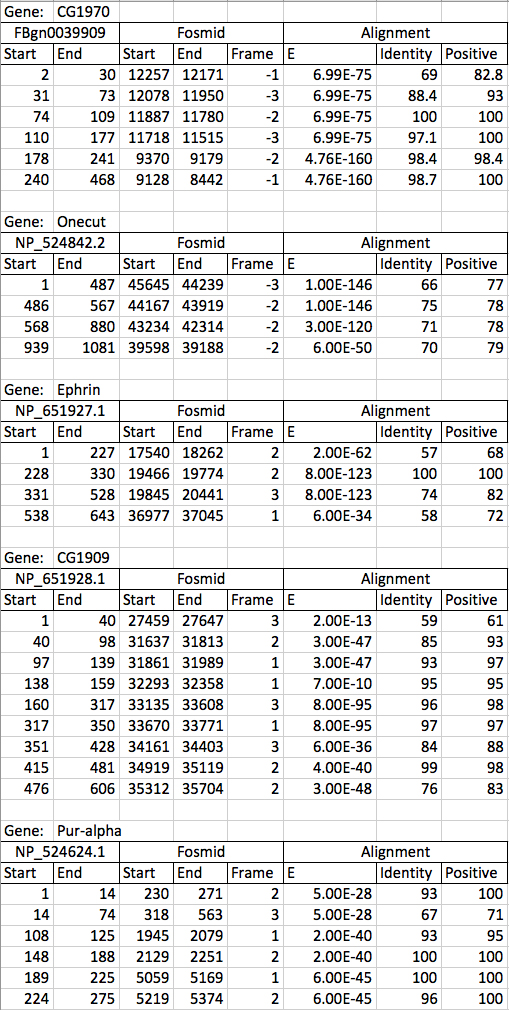
A preliminary analysis of tables indicates that all exons for all five genes are present on the fosmid though further analysis to this claim is needed. The tabulation also indicates good matching to the proteins from the named genes across the regions indicated in the fosmid.
GENSCAN Analysis:
GENSCAN analysis provided by GEP in the contig 34 folder indicates the presence of only four genes and four gene products on the fosmid. I believe this to be incorrect. The BLASTX results shown previously and the UCSC Genome Browser at GEP results later in this document indicate that the GENSCAN analysis fused the Ephrin and the CG1909 genes into one single result. I believe this to be incorrect based on the other analysis and maintain that there are five genes on contig 34.
UCSC Genome Browser at GEP:
The view of contig 34 in the UCSC Genome Browser at GEP supports the previous analysis. The browser shows good matches on multiple isoforms of the five genes. It also shows GENSCAN's error of combining the Ephrin and CG1909 genes. With the exception of the two genes Ephrin and CG1909, the gene predictors appear to strongly agree on the genes and exons in this fosmid. Conservation also appears strong across the genes.