Summary:
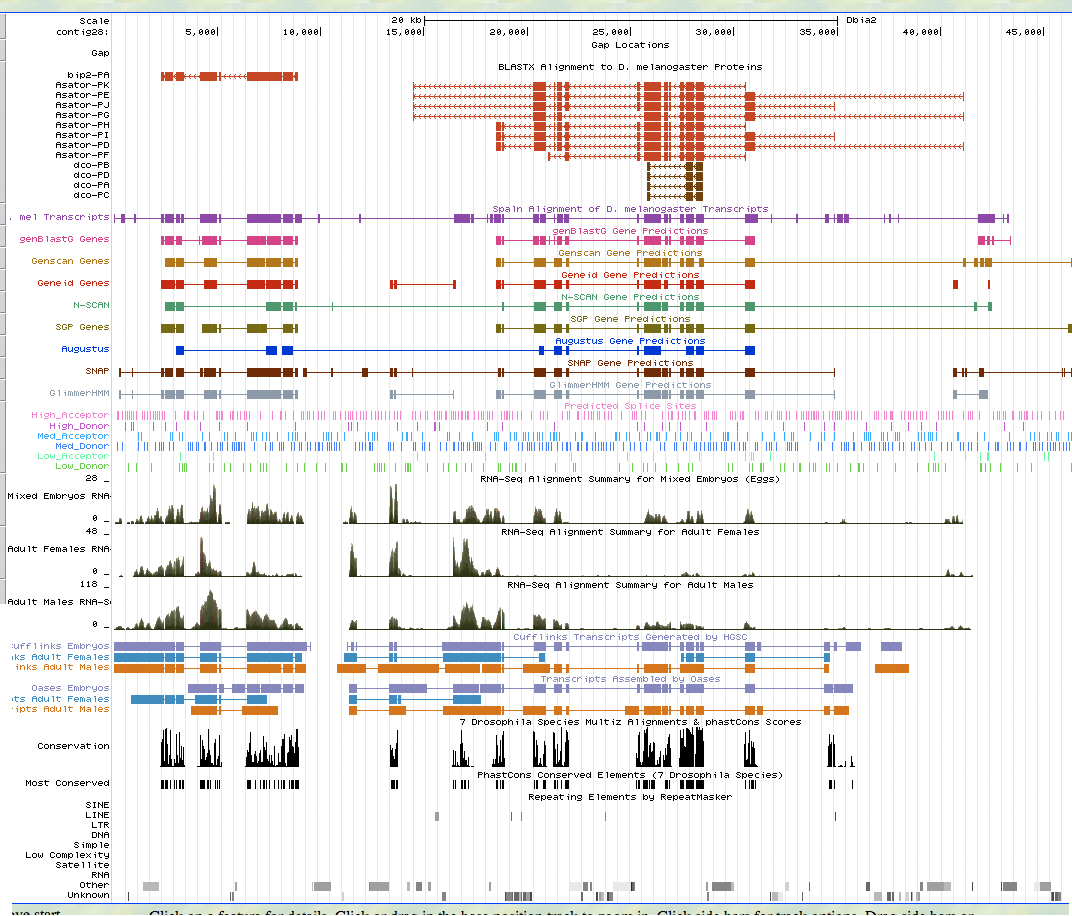
D. biarmipes contig 28 encodes two genes: BIP2 and Asator, both of which are orthologs of the D. melanogaster genes. It appears that the genes are both located in their entirety on contig 28. GENSCAN predicted two genes for contig 28; both predictions are accurate, though GENSCAN did predict numerous additional exons on the 5' end of the Asator gene.
BlastX Analysis:
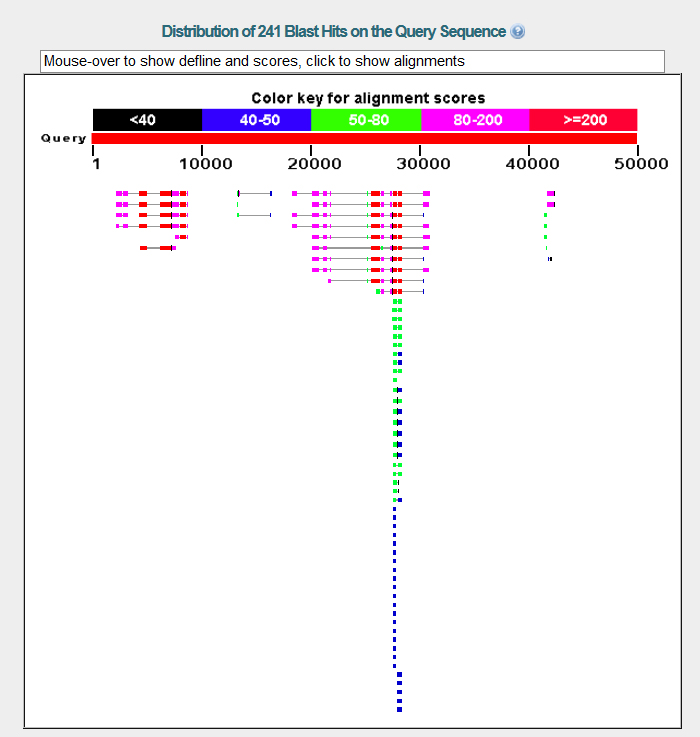
Using the given nucleotide sequence for contig 28 as a query, a BlastX search was performed against non-redundant protein sequences restricted to D. melanogaster.

NCBI BlastX found two gene matches on contig 28, both of which have significantly low E values and high identity and positive values. They appear to be homologs to their counterpart D. melanogaster genes. Other matches with high scores appear to be isoforms of these two genes, the gene on the left being BIP2 and the gene on the right being Asator.
Alignments:
Below is a spreadsheet including the exon alignments of BIP2 and Asator. The coordinates are organized by gene; all coordinates correspond to the BIP2 and Asator genes as depicted on the BlastX graphic summary.
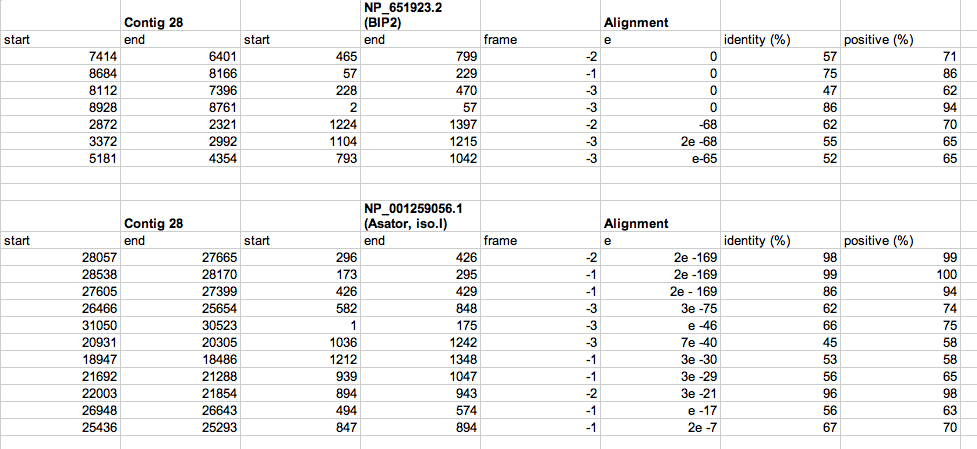
Summary of Gene 1 (BIP2):
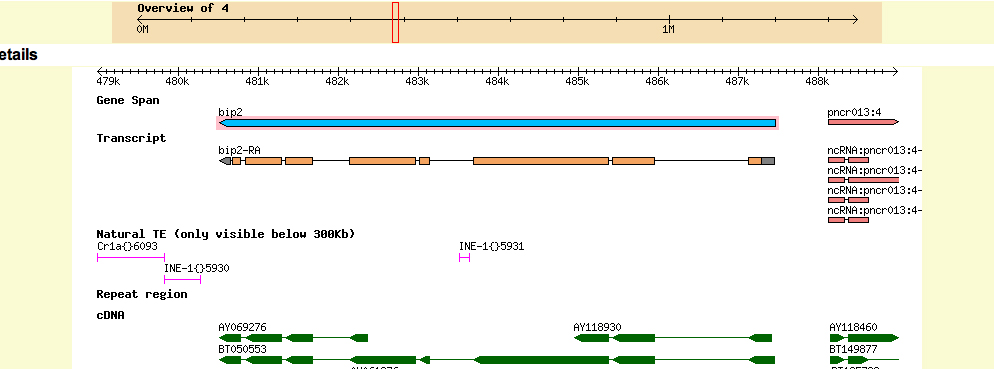
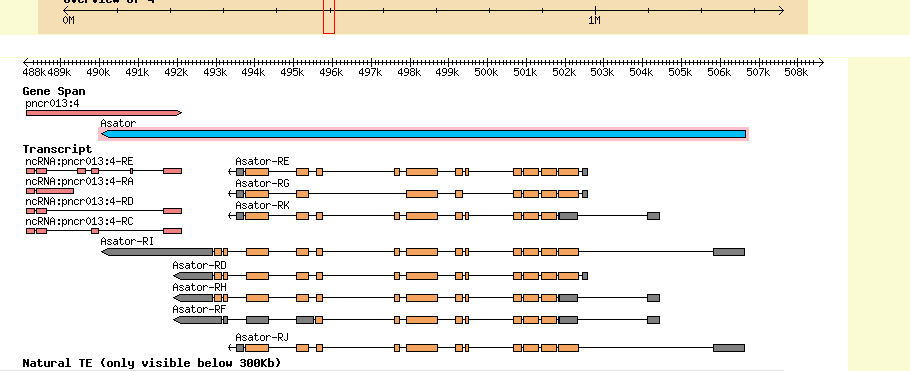
Summary of Gene 2 (Asator)
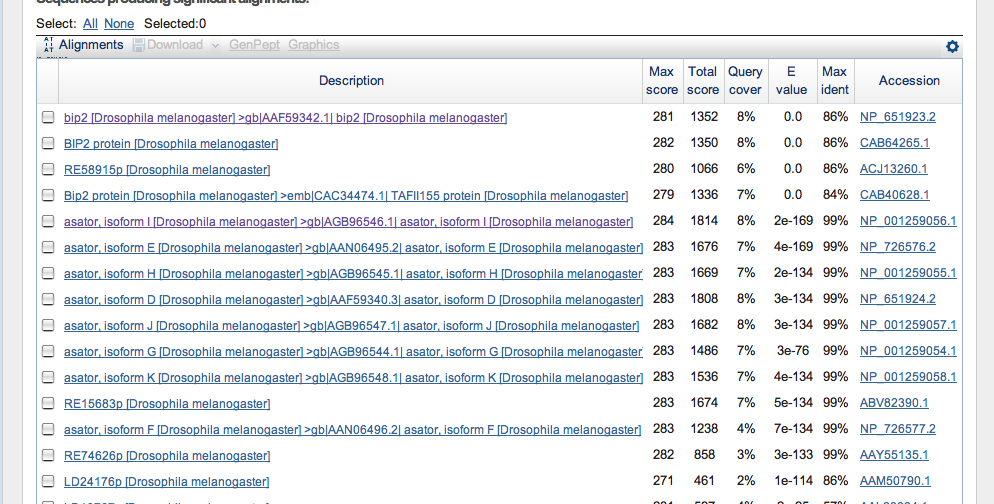
FlyBase Blast:
A FlyBase blast was also carried out for the purpose of further analysis. The database was set to annotated proteins (AA), the program was restricted to BlastX, and the organism was restricted to D. melanogaster. The graphic summary of the results is below.
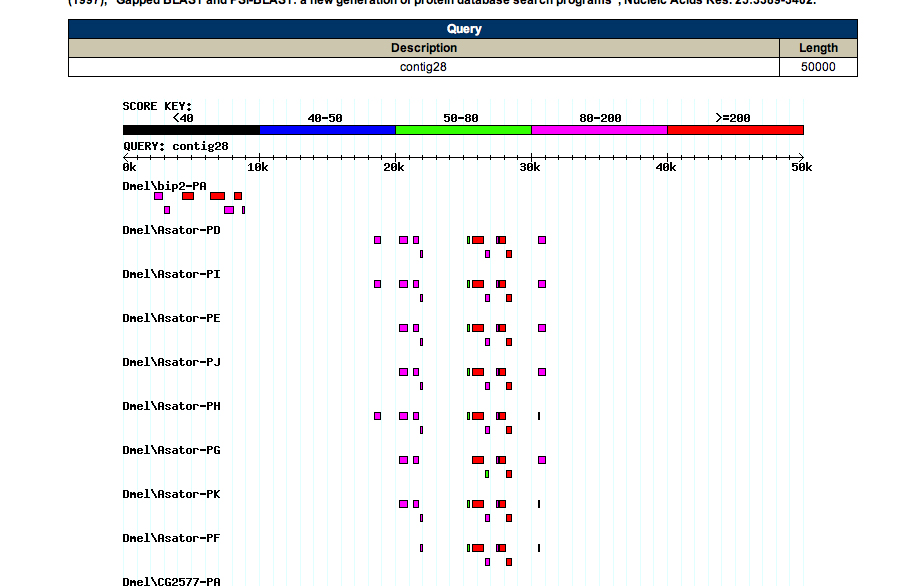
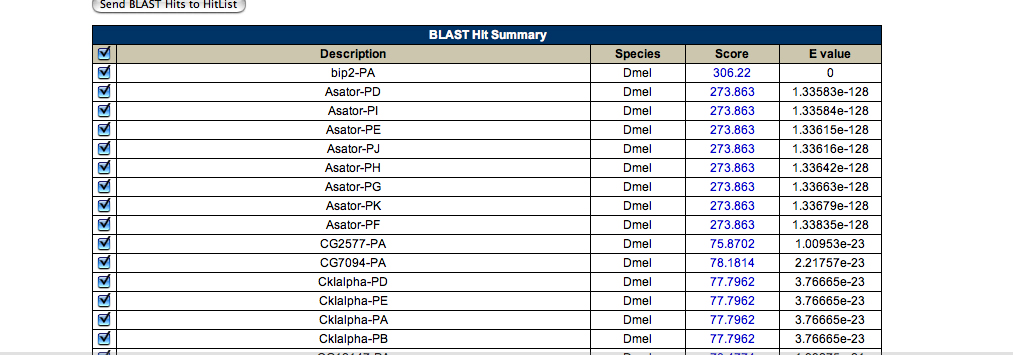
The alignment matches are as follows -
GENSCAN Analysis:
On contig 28, GENSCAN predicted two peptides as found in the project folder. The predicted proteins are as follows -
>contig28|GENSCAN_predicted_peptide_1|1273_aa MADKYASDLALVVIAQITQTIGYSCTLSAPLELLQDIMQKFTQEFARDLHCNMEHANRIE PSLRDAQLSMKNLNINIQELLDYIGNVEPVGFTRDVPHFPIRKAINMNFLKPGSAETLTR PVYIFEYLPPMQDPEPRESQSEAQREFFQKQELVSKAEFNVTSSAEKLSTNQVDSSSPNA VINFSSNFLDSDVGRSVREMSSVVMTTGGFISPAIEGKLPEPFIPDIIEKFKGLDAPPPS LIAVSHLEHSEKELITSEKDAVNTRTINSETKNFNHNAVLLISDSADASLLYSSNSLPMS SALTATISKKNRKPKPDLAIEHGQIVKSNISFGKSQEKSQRKALKMFQKLSKSQNDASNS QILNMKKSKKRVNHGNSLSDPSKVNIEKMFKKQNRHKQKSVQLETQTLSDFPIEADMNTE NFIAADIPCAEPVVLKQIEIGSQSSVFPVQPGGIQQTQVSLQSKKMHQNRNEDGGATQVP PGIGVRVFGSTMPVSTLISLPSGTTITPTPPVGQTPEDANNPNSKINQYGVLGQKLSSPK DDIEMSTINPIKAKKRGRKPGGKNLVKQTHFTSHSLIESSKKEKSTRVGAFKLASSETLV SQNSLSVSMPTEPLNLSSTDQASIEYLPNFHAKKERKKYKLKFDTGLPQNVKDFNNVFSE TTSSTVTSSIHLMKQKNEAPCPDSLMTAPNTLSNTSLYPGNQTGMVPLLPLLHFPPRPGL IPSGPGLFPAVTGLVGFGNNNNTVGIAPFIAFPGTEGSVADTVRCPPIKDSAGKPVSCGN TDMENTFTPSKFKAKPSVQASGNLGDPIEVSDDSDESIQNRQIVQKKSPLTSPNHAKSSL VQSQHQTFATPSPLCEDKNQMDLRNVLPNTSPSESIKKLKKSVKLNLPDVKNFIQIAPSQ SSFPQFNLPNFIGGDKFSLAGGADLIPLARIDCGSAYSSHKVPSSSLTGGVASGVIPILP NHISEDQQFMPTFPNYEDISITPTGALSLDPKILIQIPEVDEFSRAPFINNLDYGFTQSV PASTPVLKTSPMSVKPPLSATCAMSPKIQQSPSQIPKLTLKLSGKSTTCPEKEKDTTDAV KVSQPTMFPVENKERERDNSPELARFSPLVTGPPKNKQSDTHLLGISSAGPLLNICSGNS MKVIQEPFPMATRTSQISTSQNSSSSAGWMSNPSNSNVASSTLSASSVLLPQQLMLTSNT TMNNSLSSGGPKSCSLSSPANVPEENSHIAETNRPSSYVDAEGNRIWICPACGKVDDGSA MIGCDGCDAWYHX >contig28|GENSCAN_predicted_peptide_2|1746_aa XPTKNFGGAILVVGLGLVRVTFPTKRSSPKYQQPRRVFLSQRREPQPRAQKKLPRQNYNE VGGGAEERQSGVENRERRPARGTGPGTWALGSGLWALGTGTTSAVQLPLQQVWANDPLGK ATVFRRFYGAQAKMCQTYNEEICWNPETMVAIRKHYPFINMFSRPNWRRLLIVRDPRGVI YSRINYEWCTIKKDCKVQSLCNNMVSDPGGDIKLAFDLLGLPLGDPGPLLEGSTRALVLR RGPSVGTFVGGTSTHPLSGRVDKGCDPSIEYSKTATLYELAFISDSVFVIRIAPVNAQGP RKVAYRLGGRTLFAPLLRAVELARESGCFPPAGLGALLMGRSIDAYAVDSHTEARERGGR RAEDTDAFRGGDASPFRACFVVLGFILNDENASSPGDGNHNHTCQPPCNQEQYISLNRDC QKNLFRLHPPPPSKPPPLAGAILQSRLLKQISSGANAENAEALEEHRYPNALQRSATLPA KHNRLGVRSRVTFKVPSSITPAVDPGSEPDPGQNQVVAAERDGIVLNPKERESCKMTSED LLQPGHVVKERWKVVRKIGGGGFGEIYEGQDLITREQVALKVESARQPKQVLKMEVAVLK KLQGKEHVCRFIGCGRNDRFNYVSNFSVGRLPYNCRRVYMLDFGLARQYTTGTGEVRCPR AAAGFRGTVRYASINAHRNREMGRHDDLWSLFYMLVEFVNGQLPWRKIKDKEQVGLTKEK YDHRILLKHLPSDLKQFLEHIQSLTYADRPDYAMLIGLFERCMKRRGVKESDPYDWEKVD SATIGNISTSGNPPVPVKNDYIHGNITQMTVAASNASGTEYVRKRGDIETAHITATEPLH IKEKVDKNCNATTLAFQPKTSGEANVQLGCTANNQNITPKGMLQQQAALAINSQAAIPHM QSVPIKSPMVGMGSHDVQVHTKNSQPQKGAASFSSTNQNNSAPVYGNSYLQLEDKAPNMF LPTKPNAESESTVDVAPKSIFEEKFVDSNDAECANKAFLSGEQQQQKSQVKKLNLPESAN QQVQKLENVANEKSSEDKRASQEPKSTFGRLRVLTAPPMSVHDLTTGGHIQQGTDLSIKQ DPSSSNAGPAAGNSSSSKLAINQHGQIFGITLMPQVNRRSATSTNLRPSSSGGNTNPIHR INIGSAGGGGGTGSNTARSSVAGDHSVTQFALIDDENVSALQQVTKGGALTLASQWKSQF DDSEDTTDNEWNREPQSQPNLEQLIKLDIPLPLNEAKHFCQNVVTDTGILTKPPIEGNEK PKRYTLNITGIENYEALRISIPHCWSEPAMGNVLRKDLEPPAVQQAAFDDTVYRMDIARN VCVRETYSDITPLDKAKPATSLVSRVVLPSPFKEDATFQLNTSNNSQAKLKHRRSLPNVS VADLFDDQPIHSNSDAMLAEDANKPWKVQTMACQRSNVALSSAVQENNGCISGRLEIRVI PKETSHLDDSVYYDALAPIKNTTTANPDPGISDKANIFCDEIEENAAVIALPSNTANKCK KQTYTDKTEACIEANPCSIHATGVNNLKINGKSETCNALNDQPNYKSGDYKTPFTGGSTD SYRETDSGCDLPLLNPSKIPIRQSKCASWAGADTTVYSSKTLEPRDALPEIPFNPQTNTY STALEYPPNITDLTPGLRRRRESAEGKYVTDQTQLQLKFQRPRSRTSSRTRGISNTMLGN FDDNNTVSGEKQRLIGTQVVQNEESNNVPTSIYSAELQDKCNISPPPGDPKIENSARLRR YRHNLE
Each predicted peptide was used as a query to carry out a BlastP search of non-redundant protein sequences restricted to D. melanogaster proteins. Both proteins align significantly with D. melanogaster. Results are summarized below:
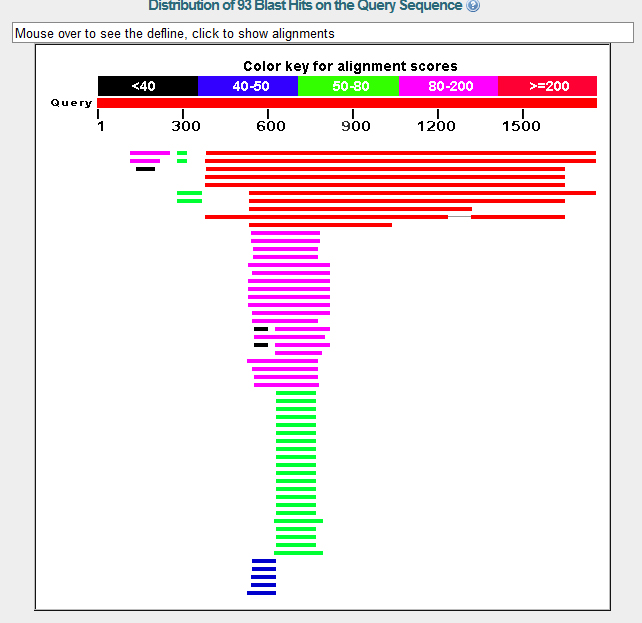
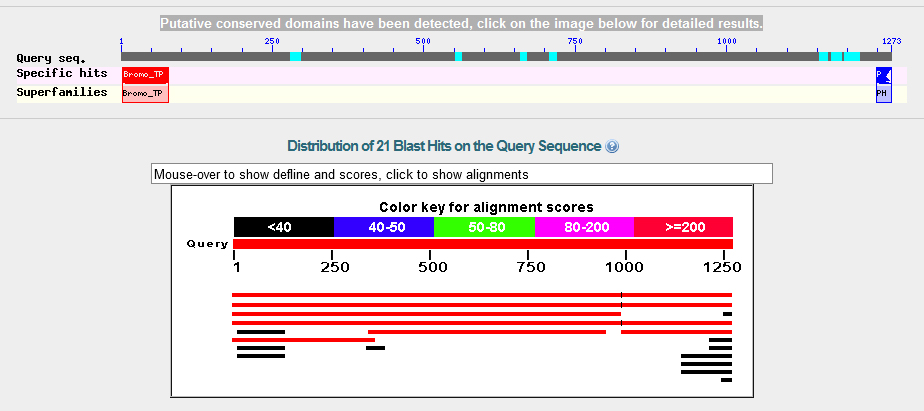
The data suggest that the GENSCAN predicted genes are accurate. GENSCAN predicted the genes BIP2 and Asator. No additional genes were predicted.
UCSC Genome Browser Analysis:
BlastX Alignment of D. melanogaster proteins:
GENSCAN predictions: As depicted, the GENSCAN predicted genes align to their D. melanogaster orthologs. However, GENSCAN predicts numerous additional exons for Asator on the 5' end.
modENCODE RNA-Seq:
Conservation: