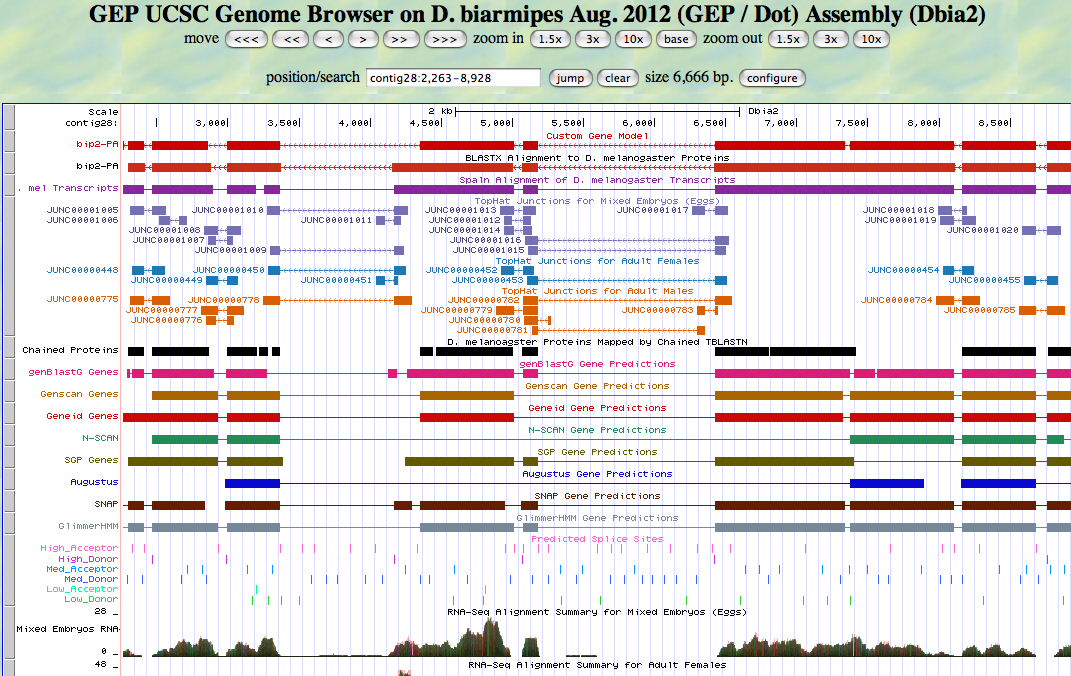
The D. biarmipes bip2 gene is located on the minus strand of contig 28.
The Gene Record Finder gives 8 CDS sequences for bip2 in D. melanogaster. Below are the exported CDS sequences.
bip2:8_1649_0 MMADRYASDLALVVVAQITQTIGYSCTLSAPLELLQDILQKFVQEFARDM HRHMEH >bip2:7_1649_2 NRIEPNLKDARLSIKNLSINVQELLDYIGNVEPVGFIRDVPQFPIGKSVN MNFLKPGSAETLTRPVYIFEYLPPMQDPELREIPADVQKEFSEKQEFCSK AEYSSTNAADKLGAKHIDSISPNTVINFRSNAFELDVGSSVREMSSVVMT TGGFISPAIEGKLPEDIIPDIV >bip2:6_1649_2 KFLGLDAPFSPTIIVNSLQKSPQLALSDRDKTVNPRKIIPSETKIFKQNA ALLTSGHSESSIIYASNNHTMLTVTTPKKNRKQKHDLICEPGQSELLTNP FEKAQEKSQRKALKMYKQTSKNQRDSSINQIQNMKKLKKKFNRGSFDPNK KHLEKIFKKQSKLKQNDLQLDIDEKHFLQNSQTTSGLAIKANSNENALQA DIPSQPPTVTSQIENNFRNSIYPVQPSVIQQTQVLLAEKKSGSEPERSKL DIFKKISKPRTPRPDIGATLVPPGTGVSVFGSTMTPSALISLPSGTTITP TPSLGLNSENKNVPSMKINPCNIFDGTIPLTKAGVEMSIIDSPKPKKRGR KPGGKNVIKQTNVVSQPLINKVERKKSSQAITLPLSSSPMIITQSSLNVS PPTEPLNLCNTEQPSNNFLSNLYAKEKKDRKKYKSLPENVMQLDKSCSPE KASTLHNSIRPTKQNNDVQCSDKFVTISNALSNASIYPGIQTGMVPLLPL LQFPPRPGLIPTGPGLFPAVTGLVGFGNHGNRIPISPFIDYPGPEESVAD TVRCPPIKDSP >bip2:5_1649_2 TDSDQFLSRSSTQTDLQMDRNYCNVAPLVPDSMKFAE >bip2:4_1649_2 KSVSCASTILENPSAQATSKINTKPLLEASGNPDDPIEVSDDSDESMHNR QMVQRKTPISSPTYVKTSFAELNPSSFTSAASNGEEKSKMDLRNIVSLSS NEPLKKFKKLVKQSFPDVKSVPHTPASHSSFPQFNLPNFMGGDKFSLAGG ADLIPLSRVSDSEYSSKIVPFSSLGGTIPNQIKISEEHNIFSTFSNYEDI TITPTGLTSLEPKMRKHHKKLKKVKEGKNKKKKEKKDKSKKADQIGLPSF KSDRKIKANDKRQKKEKKKDKDKQ >bip2:3_1649_0 ILVHIPDDTEEFDKVPLANNDEPVLKSSSMTINPSLGAATSGISPNQIPK LTLKLSGKSTLFSSSEKEMTDAGKLKQTTILSSENKKRERDNSPELARFS PLVTGPPKNKQ >bip2:2_1649_2 ETLHLGNSSTAVLPVPSPVAVRAVQLPVSQTSSNSAGWLSNPNNSNTASS TLSASSVLLPQQLMLAPHTIMNNFVPAMCNSTGTVSKSGLCSSPPNTSEE NANAMQIAESSRPSSYVDAEGNRIWICPACGKVDDGSAMIGCDGCDAWYH >bip2:1_1649_1 ICVGITFAPKDNDDWFCRVCVTKKRIHGSEKKKRRNKKK*
These peptide sequences were used to carry out BLASTX searches against contig 28. The results of these blasts are included below.

Exporting and blasting all fasta sequences for bip2 initially led to some confusion, as two different but overlapping "6" exons were found. After looking at the available evidence, I concluded that there is an additional intron in the D. biarmipes bip2 gene that is not present in the D. melanogaster ortholog.
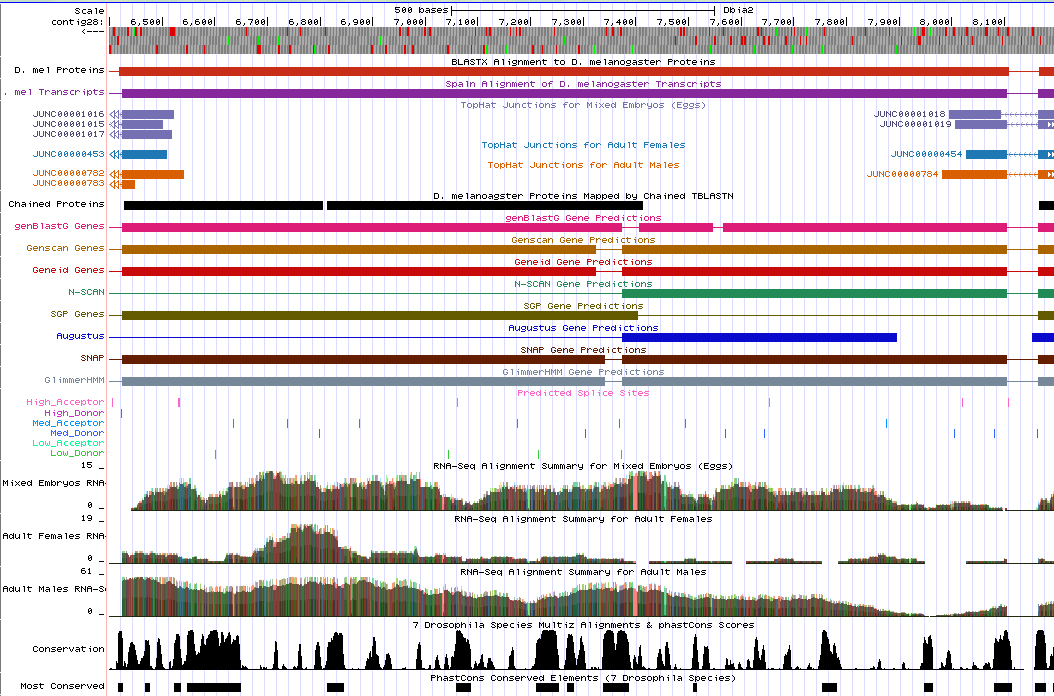
The evidence for the additional intron is as follows:
The two "6" exons cannot be one exon in D. biarmipes because they are observed in different reading frames (-2 and -3), as is shown in the above picture.
Additionally, there are appropriate splice donor and acceptor sites at the 3' and 5' ends of the two exons.
Finally, a search of the bip2 gene across various Drosophila species revealed that this additional intron is also present in the D. grimshawi species.
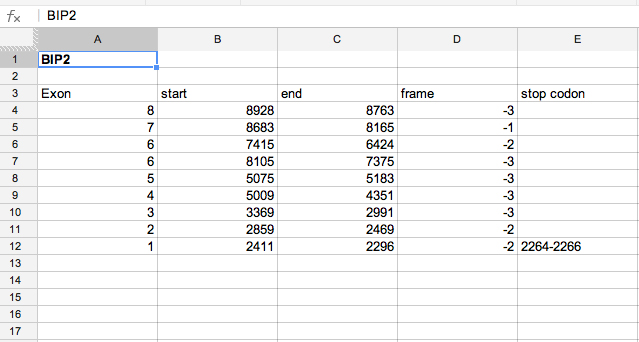
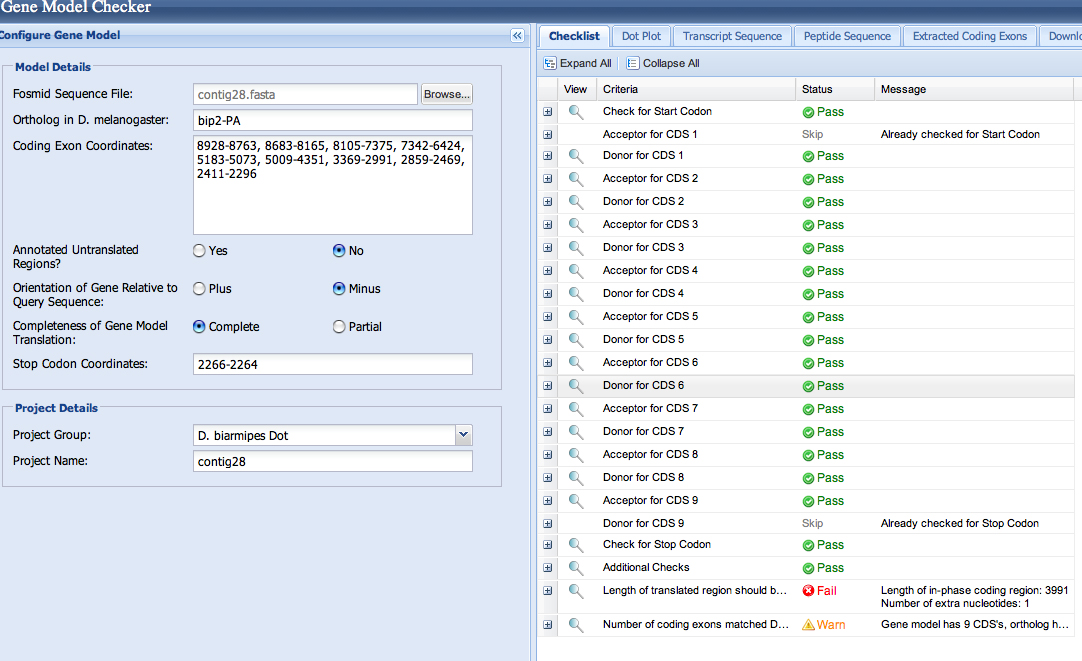
I entered the 9 exon coordinates into the Gene Model Checker to verify the gene model. The checklist indicates no issues beyond a warning about an additional CDS not found in D. melanogaster (as could be expected), as well as a warning about an additional nucleotide (acceptable with the addition of an intron).
The dot matrix alignment of the bip2 D. biarmipes gene against the D. melanogaster ortholog shows a good alignment.
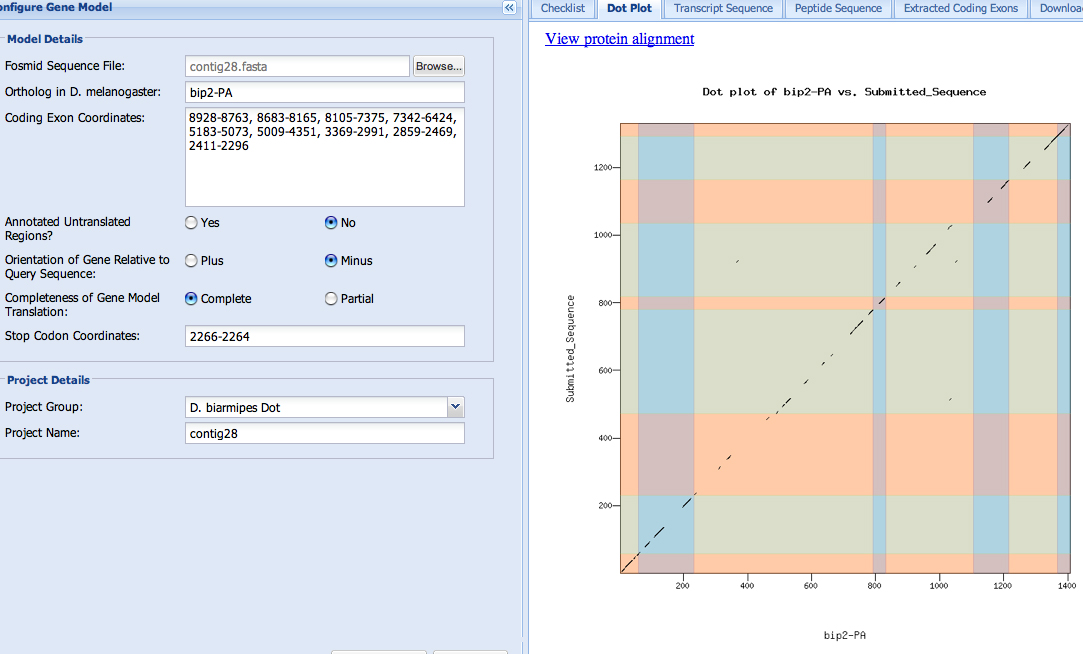
The actual alignment of the sequences also looks good.

I imported my gene model into the Genome Browser as a custom track. The custom track aligns very well with the D. melanogaster ortholog. Note the additional intron in the coordinates 8105 - 6424.