| Summary | |
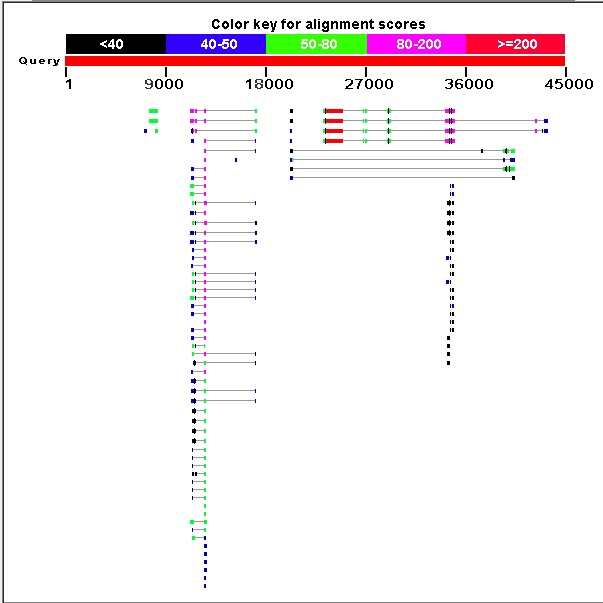
| BLASTX Analysis Graphic summary | |
| BLASTX Analysis Descriptions | |
| BLASTX Analysis RhoGAP102A alignment | |
| BLASTX Analysis dpr7 alignment | |
| BLAST Analysis by GEP | |
| BLASTX Analysis at FlyBase | |
| GENSCAN Analysis | |
| UCSC Genome Browser at GEP |
D. biarmipes contig 16 encodes two genes: the D. biarmipes orthologs of the RhoGAP102A (Gene B) and dpr7 (Gene A) D. melanogaster genes.
I used the file contig16.fasta from the src folder in the GEP project file for D. biarmipes contig as a query sequence in a BLASTX search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings.
Graphic summary. The graphic summary is shown below.

The BLASTX search suggests there are two genes on the contig with protein sequence similarity to the D. melanogaster protein set.
The left gene (Gene A) has many good matches to D. melanogaster proteins. The two best matches are different isoforms of the same gene. Other matches are still good and appear to be copies of the gene.
The right gene (Gene B) has four good matches. These appear to be isoforms derived from alternative splicing. There are many poor-scoring hits that cover only a small portion of the top hit.
Descriptions. All descriptions with E values smaller than e-10 are shown below.
Alignments
Gene B. The first four alignments in the list of descriptions all align to a region of the contig corresponding to Gene B in the graphic summary. The coordinates of the top hits are shown in the table below.
| contig | NP_001033802.2 | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 23501 | 25117 | 78 | 621 | 2 | 1E-174 | 62.68 | 75.74 |
| 23280 | 23516 | 5 | 82 | 3 | 1E-174 | 53.09 | 71.6 |
| 34875 | 35165 | 959 | 1055 | 3 | 2E-94 | 88.66 | 92.78 |
| 34322 | 34606 | 814 | 908 | 2 | 2E-94 | 66.32 | 78.95 |
| 34666 | 34824 | 907 | 959 | 1 | 2E-94 | 83.02 | 90.57 |
| 27104 | 27238 | 685 | 729 | 2 | 3E-22 | 73.33 | 88.89 |
| 26889 | 27035 | 635 | 681 | 3 | 3E-22 | 59.18 | 83.67 |
| 29203 | 29385 | 760 | 824 | 1 | 2E-19 | 58.46 | 70.77 |
| 29048 | 29152 | 727 | 761 | 2 | 2E-19 | 71.43 | 80 |
Reordering these in order of the segments of NP_001033802.2 gives the results shown below.
| contig | NP_001033802.2 | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 23280 | 23516 | 5 | 82 | 3 | 1E-174 | 53.09 | 71.6 |
| 23501 | 25117 | 78 | 621 | 2 | 1E-174 | 62.68 | 75.74 |
| 26889 | 27035 | 635 | 681 | 3 | 3E-22 | 59.18 | 83.67 |
| 27104 | 27238 | 685 | 729 | 2 | 3E-22 | 73.33 | 88.89 |
| 29048 | 29152 | 727 | 761 | 2 | 2E-19 | 71.43 | 80 |
| 29203 | 29385 | 760 | 824 | 1 | 2E-19 | 58.46 | 70.77 |
| 34322 | 34606 | 814 | 908 | 2 | 2E-94 | 66.32 | 78.95 |
| 34666 | 34824 | 907 | 959 | 1 | 2E-94 | 83.02 | 90.57 |
| 34875 | 35165 | 959 | 1055 | 3 | 2E-94 | 88.66 | 92.78 |
Summary of Gene B: Gene B appears to be the D. biarmipes ortholog of the D. melanogaster gene RhoGAP102A. The gene is on the plus strand (all matching reading frames are 1, 2, or 3). The entire gene appears to be on the contig. A view of D. melanogaster RhoGAP102A from FlyBase GBrowse is shown below.
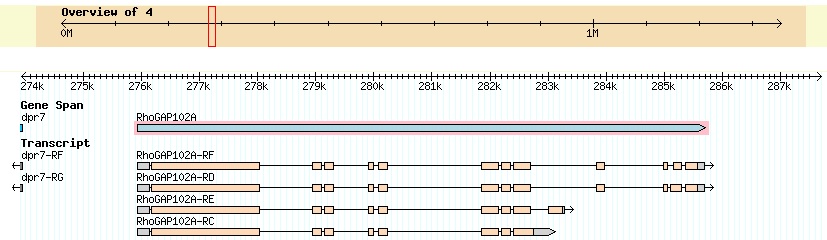
The D. melanogaster RhoGAP102A gene has four isoforms derived from alternative splicing in the 3' of the gene.
To confirm this, I used the Gene Record Finder at GEP. This returns the following protein sequences for the exons of D. melanogaster RhoGAP102A:
>RhoGAP102A:1_1653_0 MEFPKENEESVNCLRSPMAAVKKRSGPLDIALDEANDVSKIWSLPLGNAS SMRKPMIFASEHSEGVASPNKVRRRSTKDKEKKRERWLLTRKTWRYMTDA GRKLIPDGYQTGSGNHDLIEDQFQRVCLSEPSFILWSRRTSYPGAFSCSK RRLKPLLRHASASRRKQTVDATKDYHIADRVIELLQTYLKLRDAYKTTTL LGTKTVRPDQTSSPSAQRPNFKNNGDNHQVISSGTSIQTELLSLLKLLSN CPLFTHEGIQLKFGDAPLSILEDKVLLKKIYSALKKQQLHRTLHSKDPYQ ANFSTSLSCLIKNGKELSRGNTDSNRSNKLLHDNFFCLMKCDTKPIIPSP KTEFFLDLNKVPQNFELKNKLNSNGQSVERIQKTCGTQTNFIQLSELKTL AEQYNLMVTNCDNTLKLLEEPSKQDCSKSSHLIFRRSSLDEDISQSVSDT IKRYLNMARKKSMQDSDSNRFKSINYDKNLRNIKAKGVLNPPGISAGLHK AVQTLNAWPLIALDFIRGNESSLNLKNAHLEWLRLEDERSHMQLERDKNP IELRSKVNPLQIASPANTSSYSKCTSAPTSPTSHSKLEKAIRTSSGLLSS SSQFISNILHGHNNAGNQFNTLTDKLPDC >RhoGAP102A:2_1653_2 QSHATASMQKSKSLSNVGQFVAKRMWRSRSKSQNKRSYLKSSIPTLKWYP S >RhoGAP102A:3_1653_0 EHFLWISESGESFQIVETNLTRLSKIESDIVKNFALEKIQELNIGNIE >RhoGAP102A:4_1653_2 LKISPQKRRCVPKKKSLTTSFFDNGKKDDQ >RhoGAP102A:5_1653_2 PRSVLFGTSLECCLERDSKRNVTMEDRSKYSLLSMFRGSGSTPGSVLKLN DN >RhoGAP102A:6_1653_0 VRSCESLPSKSLEYGSTELSAYVRGSFSNISKPLASLSQSENEDTELNFV KAYQEHSQLMVPIFVNNCIDYLEDNGLQQVGLFRVSTSKKRVKQ >RhoGAP102A:7_1653_0 LREEFDKDIYFGISVDTCPHDVATLLKEFLRDLPEPLLCNTLYLTFLKTQ >RhoGAP102A:9_1653_2 IRNRRLQLEAISHLIRLLPIPHRDTLYVLLVFLAKVAAHSDDIWSTEGCC LTLGNKMDSYNLATVFAPNILRSTHLTFSRDKEQENMIDAINVVRFVKQN TYLLFMPLIRPNLPSR* >RhoGAP102A:8_1653_2 IRNRRLQLEAISHLIRLLPIPHRDTLYVLLVFLAKVAAHSDDIWSTEGCC LTLGNKMDSYNLATVFAPNILRSTHLTFSRDKEQENMIDAINVV >RhoGAP102A:10_1653_1 RRGSKGTEEPQPKRKWISRPGPDAQQKSPVMLLPHASYKKYSDILSLRLR QELKLERTNLERRAFIHINNKECNGLGSRPADTR* >RhoGAP102A:11_1653_1 TMINHYEEIFNISAELLNVIYTQALEACPEKLYELISTKVYGTE >RhoGAP102A:12_1653_1 TQQQIDDPQPGSLSDVLLEPPIHGK >RhoGAP102A:13_1653_1 YENINDFNNINFKRCQDKRRDHSDPDKKKNNNENLEIITASLKISVAEQS HISLKEPIKEQ >RhoGAP102A:14_1653_1 CQDKRRDHSDPDKKKNNNENLEIITASLKISVAEQSHISLKEPIKEQ >RhoGAP102A:15_1653_2 PITSYKQFSRSTLPTSISDVGVNTLRTDGAKLENKMKILTSNINKQDIPI KTTYKRQNLISSSRRISQEP*
These sequences were used as subject sequences in a bl2seq search with BLASTX using contig16.fasta as the query sequence.
The RhoGAP102A gene of D. melanogaster is approximately 10 kb, all of which appears to be on the contig. The first exon was a good match (E = 1E-179) to the 23,501 - 25,117 portion of the contig in frame +2. An additional match was also found in this region, overlapping the first exon in a different reading frame (+3, E = 6E-17, contig16 23,280 - 23,516), but was rejected due to being a poorer match on the same strand in a different frame. In addition, the first exon of RhoGAP102A in D. melanogaster is approximately 2 kb, as is the better match to the first exon. The worse match is only 0.3 kb long. The second exon has only one good match (E = 2E-14) from contig 26,883 - 27,035. The third exon has one good match from contig 27,095 - 27,238 with E = 7e-18. Exon four of RhoGAP102A matches 29,060 - 29,149(E = 7E-10).
Alignments
Gene A. The fifth and sixth alignments in the list of descriptions align to a region of the contig corresponding to Gene A in the graphic summary. The coordinates of the top hits are shown in the table below.
| contig | NP_001096850.2 | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 11713 | 11327 | 202 | 312 | -3 | 3E-67 | 74.42 | 81.4 |
| 11985 | 11752 | 149 | 206 | -1 | 3E-67 | 61.54 | 67.95 |
| 12808 | 12602 | 83 | 151 | -3 | 6E-37 | 92.75 | 95.65 |
| 17337 | 17212 | 42 | 83 | -1 | 3E-13 | 83.33 | 95.24 |
Reordering these in order of the segments of NP_001096850.2 gives the results shown below.
| contig | NP_001096850.2 | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 17337 | 17212 | 42 | 83 | -1 | 3E-13 | 83.33 | 95.24 |
| 12808 | 12602 | 83 | 151 | -3 | 6E-37 | 92.75 | 95.65 |
| 11985 | 11752 | 149 | 206 | -1 | 3E-67 | 61.54 | 67.95 |
| 11713 | 11327 | 202 | 312 | -3 | 3E-67 | 74.42 | 81.4 |
| Dmel gene | Representative protein sequence | contig | E | Conclusion | ||
| start | end | strand | ||||
| RhoGAP102A | NP_001033802.2 | 23501 | 29152 | plus | 6e-179 | Dbia RhoGAP102A (see above) |
| dpr7 | NP_001096850.2 | 11713 | 17212 | minus | 4e-72 | Dbia dpr7 (see above) |
I used the link to FlyBase BLAST from the Tools page of the course website.
I set the Database to Annotated proteins (AA), the Program to BLASTX, and uploaded the fosmid sequence. I restricted the species to D. melanogaster and clicked BLAST.
The graphic output is shown below. Notice that each hit is labeled, unlike the results at NCBI BLAST.
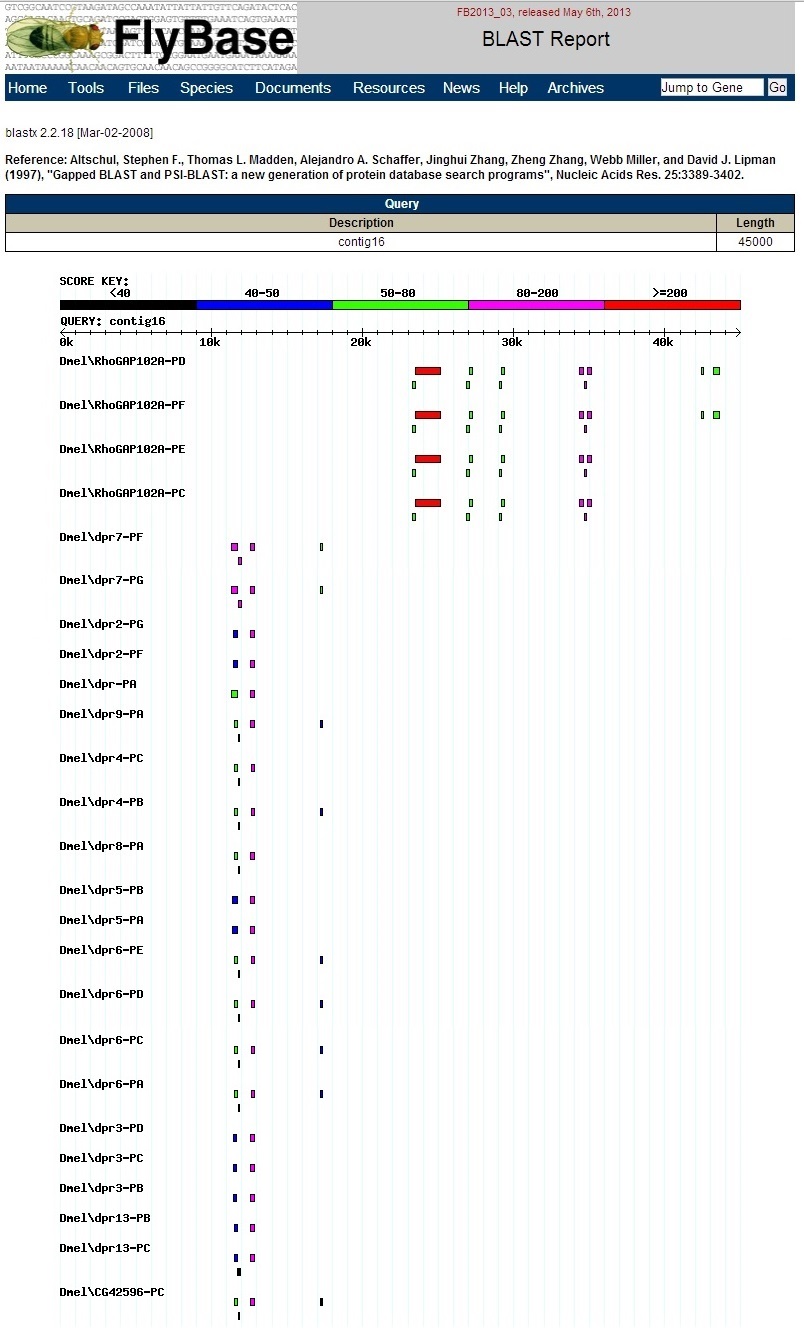
A summary table is also shown below.
The GENSCAN results from analysis/Genefinder/Genscan in the project folder predict four proteins on the contig:
>contig16|GENSCAN_predicted_peptide_1|210_aa MPRGNNATYSDNSSIKIKPFIILILNFNICLQQINASSFLNFNDLTSSDKPYFDDISPRN VSAVVDEIAILRCRVKNKGNRTVSWMRKRDLHILTTNIYTYTGDQRFSVIHPPSSEDWDL KIDYAQPRDSGVYECQVNTEPKINLPIVLEITDFDSLRGGISLETEKTEIGTTSRLMLTR ASLRDSGNYTCVPNGAIPASVRVHVLTGKH >contig16|GENSCAN_predicted_peptide_2|929_aa MTDAGRKLIPDSYPAGSDNFDLLEEHFQRVCLSEPSFILWNRRTSYPGAINSSRRRLKQL SRHACSSHKENSIETKNSYYNADRTIELLQTFLKLRDAYKTTTLLGTKTVRPDQTSSPSA QRPIFGKKSEKDQTMSSDMTLQKELICRLKLLSSCINFAQVGIKLEISDLSETILEDKVL LKKIYSALKKQQLHRTLHSKEPNKKSVNRSSSLSSLKIGEHESMSSKTSEDQKLKNQNFQ CVKICDTSSKNSINLDLNCVIKSIEFPNKCLLSDQKIERSVKTCGTQTSFIQLSELKSLA EQYKCMVQNCDNNLQVFQEFDKQDGLTSSRSTCRKSSIDEDISQSVSDTIKRYLKMARKK SVQGSDSNRFKSVNYDQNLKNIKAKGEINPPALNDGLNKAVQTLDAWPVIALDFIKGNES SIYLQNAHLEWIRSEDEREQKQLEWNKKQKQIDKEEHTPHEINRGNASHYSTCTSAPTSP TSHSKLEKAIRTSSGLLSSSSQFISSILHGHSSAGSQYSNLGNDSVNMQKSKSLSNVGQF VSKKIWGSRFKSQSKRNFSKGLKDLPSVKWHPSDNCIWISEDGERFQIVDTLLIRLSKRE TDLVKDFALEKIEELNIGNIDDLKKTSKKRRIAPKKKSLTTSFFDIGKKDDQNERVALFG TSLECCLARDRKGSANIEDRSEHYVFRKSGSNPGSVMKLNDNVRSCESLPSKSLEFGYMD SSDCSSGSFNTIPKPAASLTQFEIGDTEPSFYKTYQDQLILMVPMFIINCIEYLEENGLQ KVGLFRVSTSKKRVKQPFCNIRGEGCVRTDRRVVSYGNHTPIYIKLYQPNSFGKQKGWHT SLYVDFRKLNSMVDEQRYSRFIHECYSADTVESCLQKMALVLNTARAFGLQNKCNFLQTQ ILFLVRNIEKGNLWPGEDKTAAVSKFSNT >contig16|GENSCAN_predicted_peptide_3|72_aa MINHYEEIFKISAELLDVIYTRVMEACPEQLYELISMKLNGYEWNLNQLDDPQPSSLGDV MFEPAVQEKRFV >contig16|GENSCAN_predicted_peptide_4|51_aa PSKEDLTAVSHKPPQHNLRPPARLSREDTGMARKPSNPCYTNLILEGVISE
| Query | Top hit | E | Coverage | Max identity | |
| Accession | Gene | ||||
| GENSCAN_predicted_peptide_1 | NP_001096850.2 | dpr7 | 100% | 79% | |
| GENSCAN_predicted_peptide_2 | NP_001033802.2 | RhoGAP102A | 0.0 | 85% | 63% |
| GENSCAN_predicted_peptide_3 | NP_001245416.1 | RhoGAP102A | 3e-28 | 98% | 68% |
| GENSCAN_predicted_peptide_4 | NP_649295.1 | CG9389 | 1.8 | 88% | 24% |
Because no significant matches were found with peptide 4, I repeated the BLASTP search of nr with the species restriction turned off. Again, no significant similarities were found.
Summary of GENSCAN analysis: GENSCAN identified three genes found by the BLASTX search (Hem, Aats-ile, and Ten-m). It made two additional predictions that are invalid.
Here is a view of contig16 in the UCSC Genome Browser at GEP.
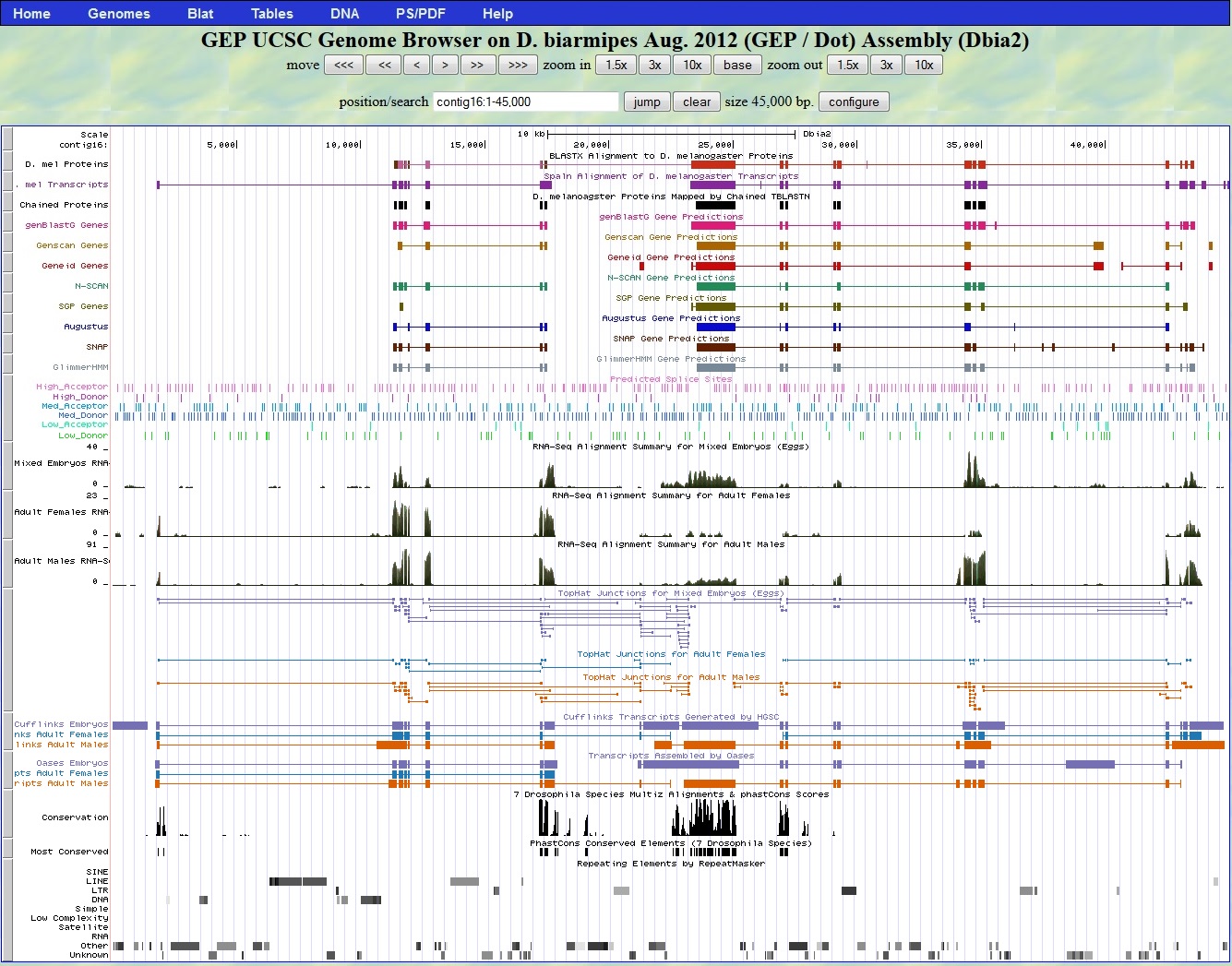
BLASTX Alignment of D. melanogaster proteins. The BLASTX track at the top of the image shows alignments to two overlapping regions, as was seen in the prior BLASTX analaysis. The left D. biarmipes gene, dpr7, aligns to the protein product of the D. melanogaster dpr7 gene and many homologs of this gene. The right D. biarmipes gene, RhoGAP102A, aligns to the D. melanogaster RhoGAP102A gene. These are the same results seen when the contig is used as a query sequence in a BLASTX search of D. melanogaster proteins. There are two genes on the contig: the D. biarmipes orthologs of dpr7 and RhoGAP102A.
GENSCAN predictions. Starting at the left of the contig, the first three GENSCAN predictions align to the D. biarmipes orthologs of dpr7 and RhoGAP102A. The fourth GENSCAN prediction does not align to sequences predicted by BLASTX analysis to encode proteins. The first GENSCAN prediction is congruent with predictions from other gene-finding programs, while the second and third predictions split the RhoGAP102A gene.
modENCODE RNA-Seq. Transcripts aligning to dpr7 and RhoGAP102A are seen. In addition there are transcripts in the 5' end of the contig at approximately 1 kb. These transcripts are a portion of the dpr7 gene that was not detected in any other gene-finding analysis.
Conservation. ??????????????????????Need to fix this part??????????????????The exons of Hem, Aats-ile, and Ten-m are clearly conserved. The intergenic regions between Hem and Aats-ile, and between Aats-ile and Ten-m are not conserved. There is considerable sequence conservation upstream of the rightmost (5') exon in Ten-m; this region is known to be a Ten-m intron, separating the third exon of Ten-m from the second exon, which is not on the fosmid.