Student name: Joseph DeAguero
Student email: josephd@unm.edu
Faculty Advisor: Dr. Paul Sauter
College/University: University of New Mexico
Project details
Project name: fosmid_2742C10
Project species: D. ananassae
Date of submission:
Size of project in base pairs:
Number of genes in project: 0
Does this report cover all genes and all isoforms or is it a partial report?
No, it is a full report.
If this is a partial report because different students are working on different regions of this sequence, please report the region of the project covered by this report: from base NA to base NA
Instructions for project with no genes
If you believe that the project does not contain any genes, please provide the following evidence to support your conclusions:
1. Perform a BLASTX search of the entire contig sequence against the non--?redundant (nr) protein database. Provide an explanation for any significant (E--?value < 1e--?5) hits to known genes in the nr database as to why they do not correspond to real genes in the project.
Summary
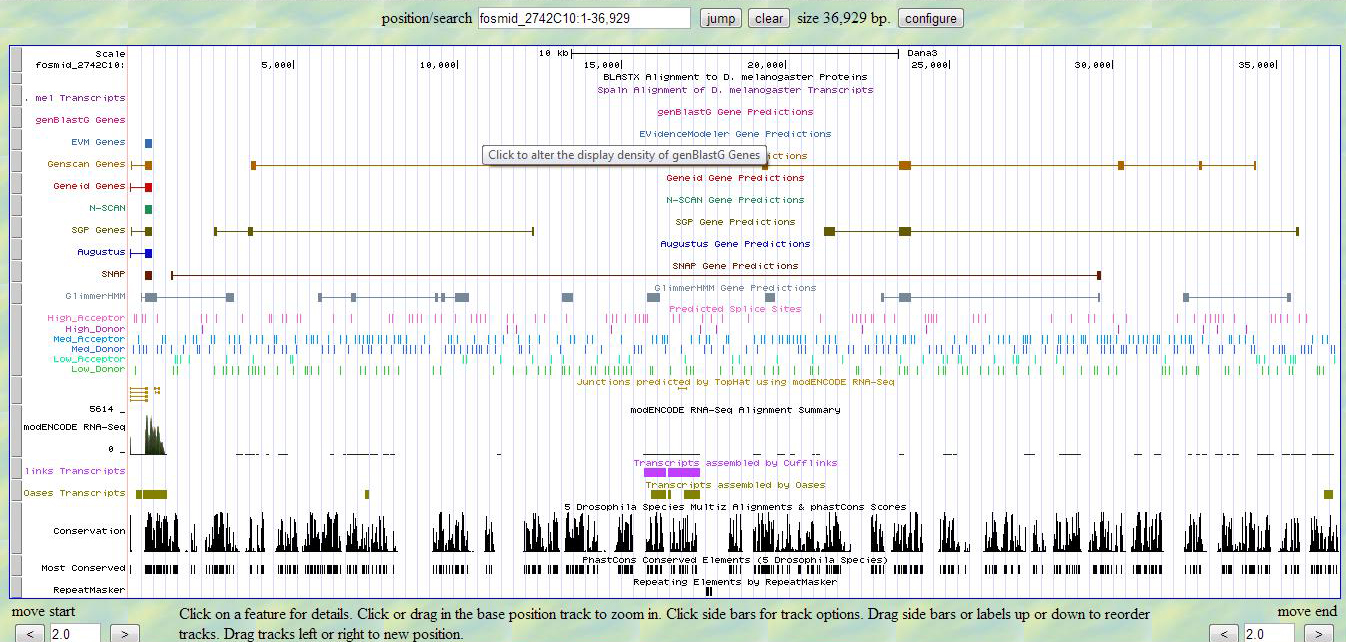
I used the file fosmid_2742C10.fasta from the src folder in the GEP project file for D. ananassae fosmid 2742C10 as a query sequence in a BLASTX search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings.

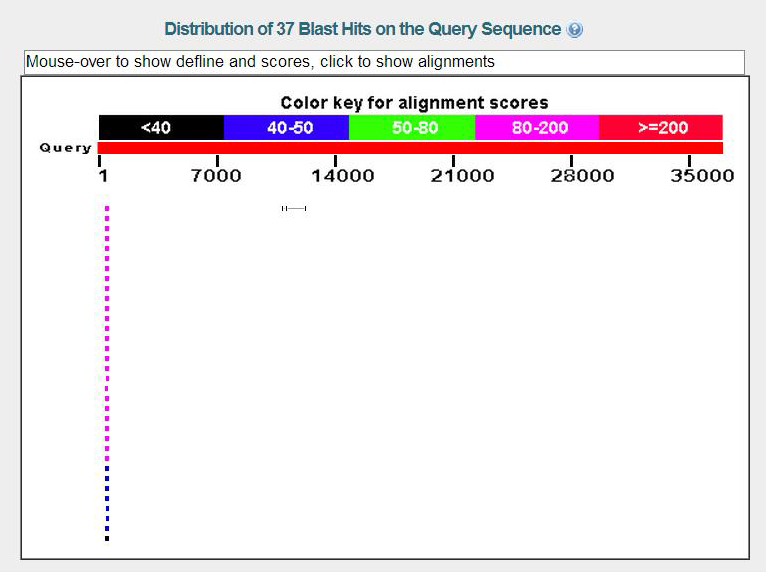
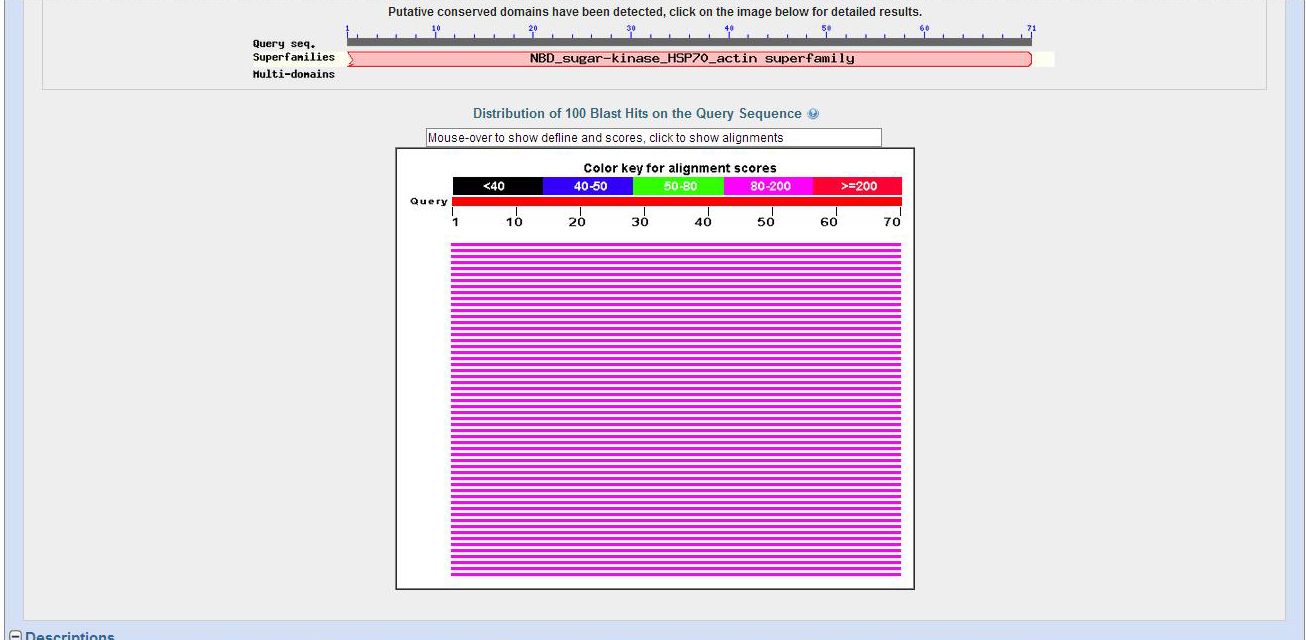
From the image we see there is a small chunk of a protein sequence in the query position 465 to 674, but there are several different Actin genes which are in this position with good E scores.
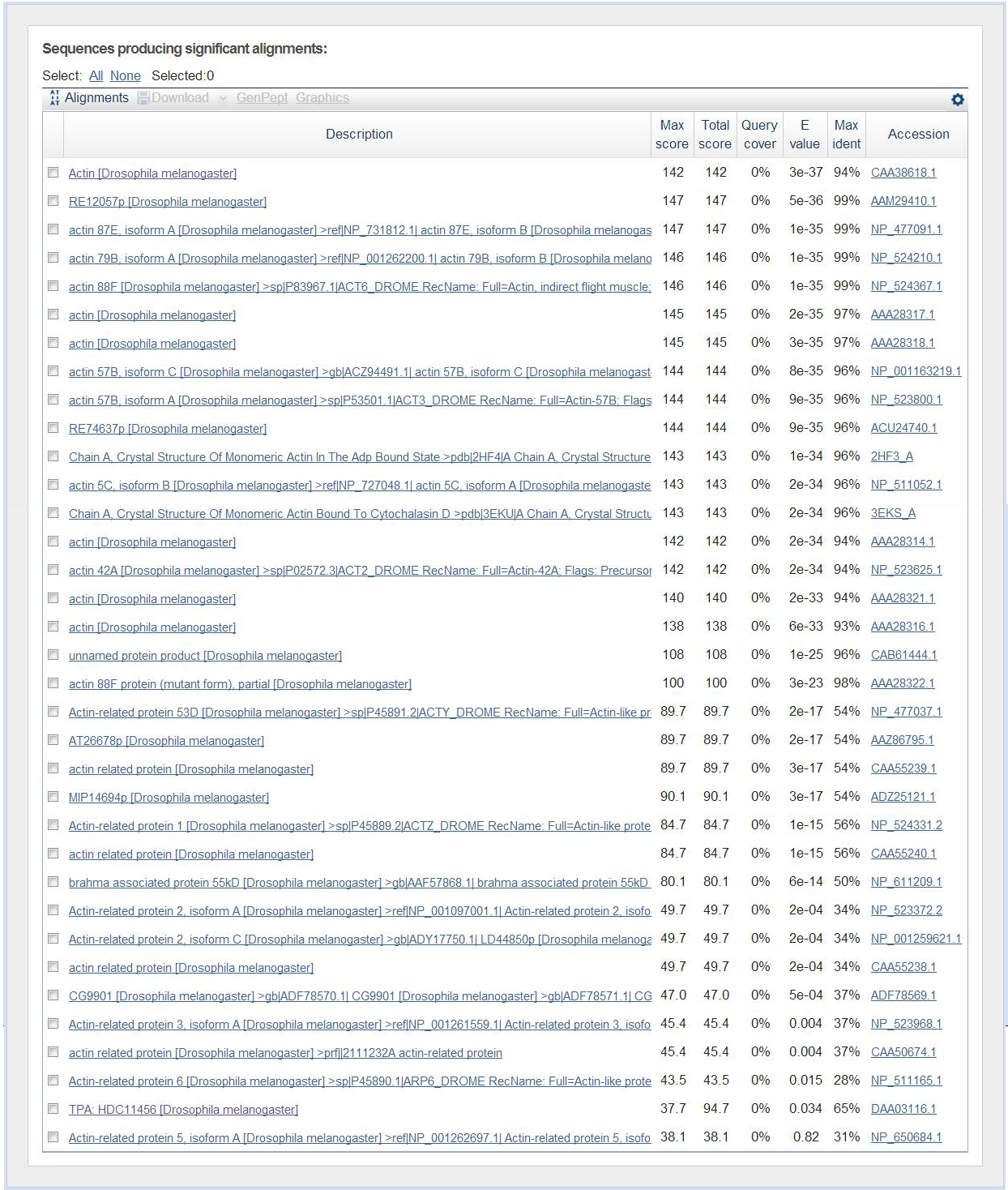
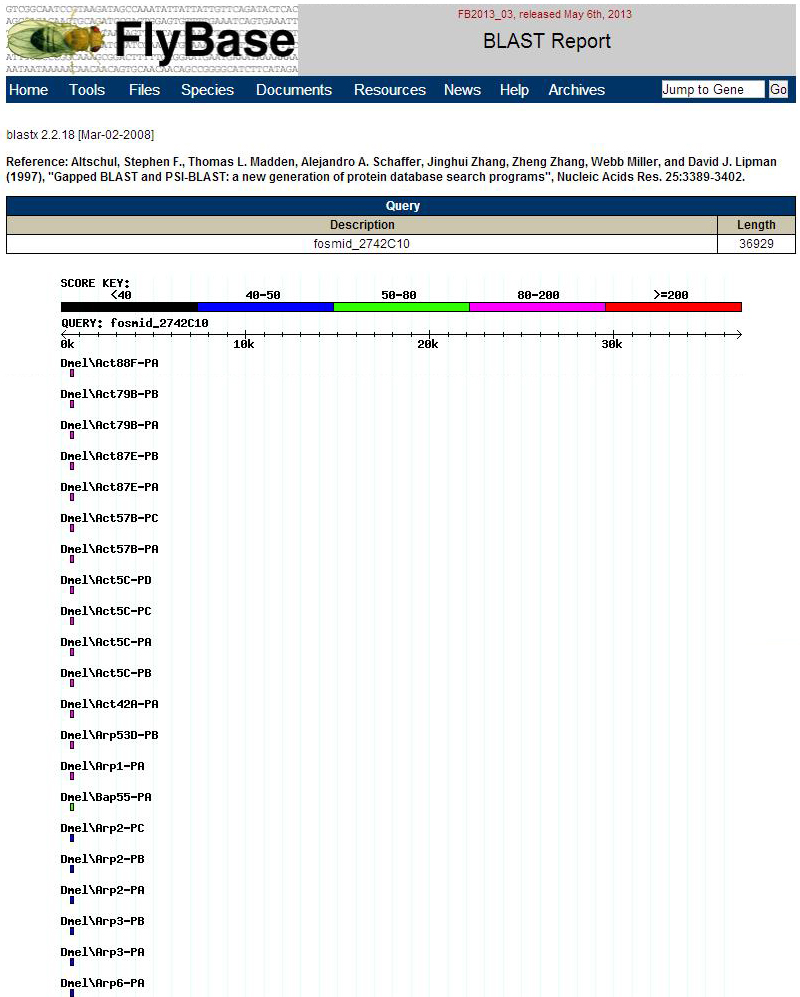
BLAST Hit Summary Description Species Score E value Act88F-PA Dmel 144.05 8.95435e-33 Act79B-PB Dmel 144.05 8.95435e-33 Act79B-PA Dmel 144.05 8.95435e-33 Act87E-PB Dmel 144.05 8.95435e-33 Act87E-PA Dmel 144.05 8.95435e-33 Act57B-PC Dmel 141.739 4.444e-32 Act57B-PA Dmel 141.739 4.444e-32 Act5C-PD Dmel 140.584 9.90019e-32 Act5C-PC Dmel 140.584 9.90019e-32 Act5C-PA Dmel 140.584 9.90019e-32 Act5C-PB Dmel 140.584 9.90019e-32 Act42A-PA Dmel 140.198 1.29301e-31 Arp53D-PB Dmel 85.8853 2.89391e-15 Arp1-PA Dmel 82.4185 3.19959e-14 Bap55-PA Dmel 76.6406 1.75567e-12 Arp2-PC Dmel 48.1358 0.000668704 Arp2-PB Dmel 48.1358 0.000668704 Arp2-PA Dmel 48.1358 0.000668704 Arp3-PB Dmel 45.4394 0.00433441 Arp3-PA Dmel 45.4394 0.00433441 Arp6-PA Dmel 43.1282 0.0215115
From the table there are several Action proteins that have good E scores but from Blastx.jpg this is a very small region.
2. For each Genscan prediction, perform a BLASTP search using the predicted amino acid sequence against the protein database (nr) using the strategy described above.
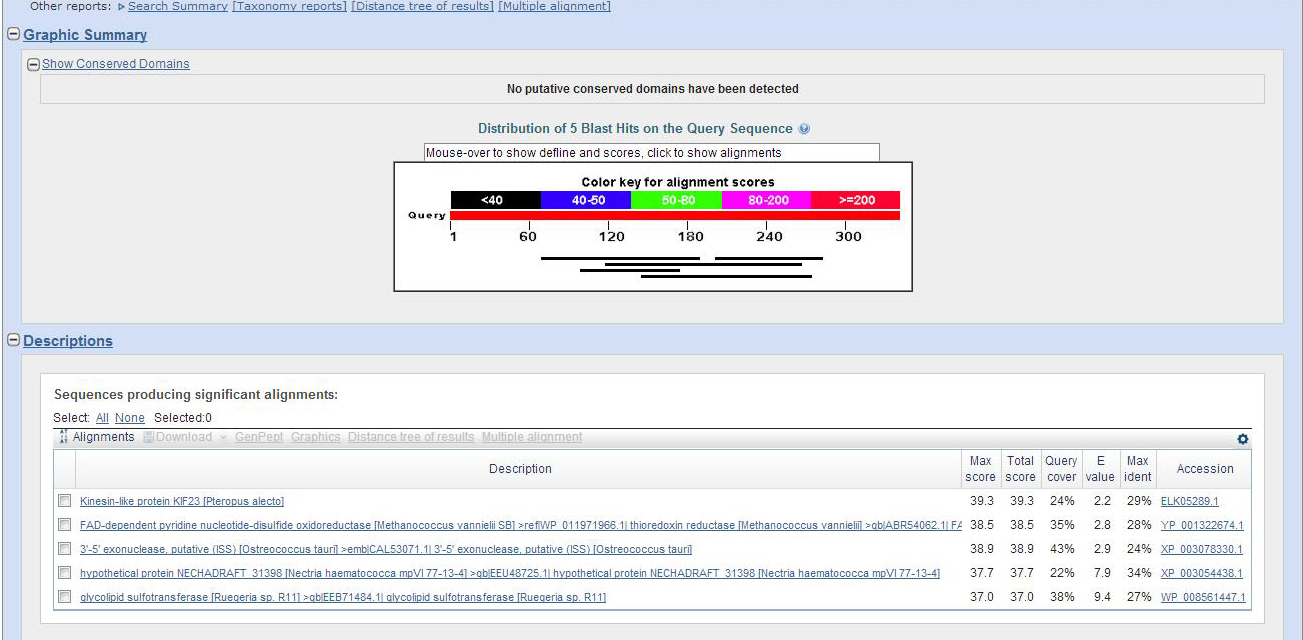
Using the predicted peptide sequences in the GENSCAN folder, we find two predicted peptides from GENSCAN. The first peptide finds Actin for all different kinds of species. The second does not find anything.
3. Examine the gene expression tracks (e.g. cDNA/EST/RNA--?Seq) for evidence of transcribed regions that do not correspond to alignments to known D. melanogaster proteins. Perform a BLASTX search against the nr database using these genomic regions to determine if the region is similar to any known or predicted proteins in the nr database.
Even though we find a small piece of Actin when looking at Looking at GenomeBrowser.jpg we actually do not find a protein.