My project is on D. biarmipes Contig 6. So far I have determined that the entire Ankyrin gene lies within my contig and that there are 4 isoforms from Ankyrin. I have also determined that though many of my alignments give me the Ankyrin2 gene, that I'm really looking for Ank because it is on the Dot Chromosome and Ank2 is on 3L.
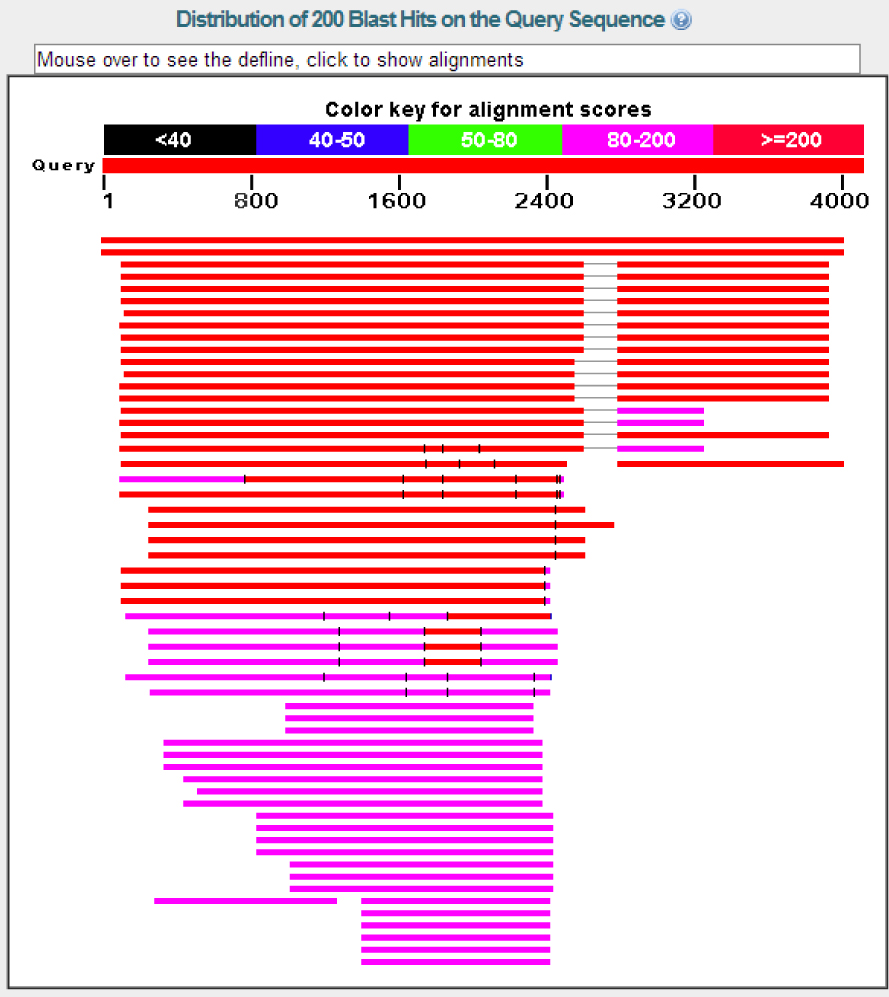
Figure 1: Here is the graphic summary of my BLASTx. I used the file for Contig 6 on the lab computers, I used the masked copy in the src folder. My contig is the query sequence and I did the Non-redundant protein sequences (nr) serach and restricted it to D. melanogaster Taxid: 7227. As you'll see in the next image, two alignments are very strong then there are many other great alignments after that. The first two alignments are Ankyrin and the subsequent ones are Ankyrin 2. For the reasons I mentioned above, I have been ignoring the Ankyrin 2 results in all my work.

Figure 2: Here are the descriptions of the alignments. As you can see I mentioned in Figure 1 that the first 2, best matching alignments are Ankyrin (one the gene as a whole and the second a specific isoform of Ankyrin (one of four isoforms)). I disregarded the Ankyrin 2 alignments.
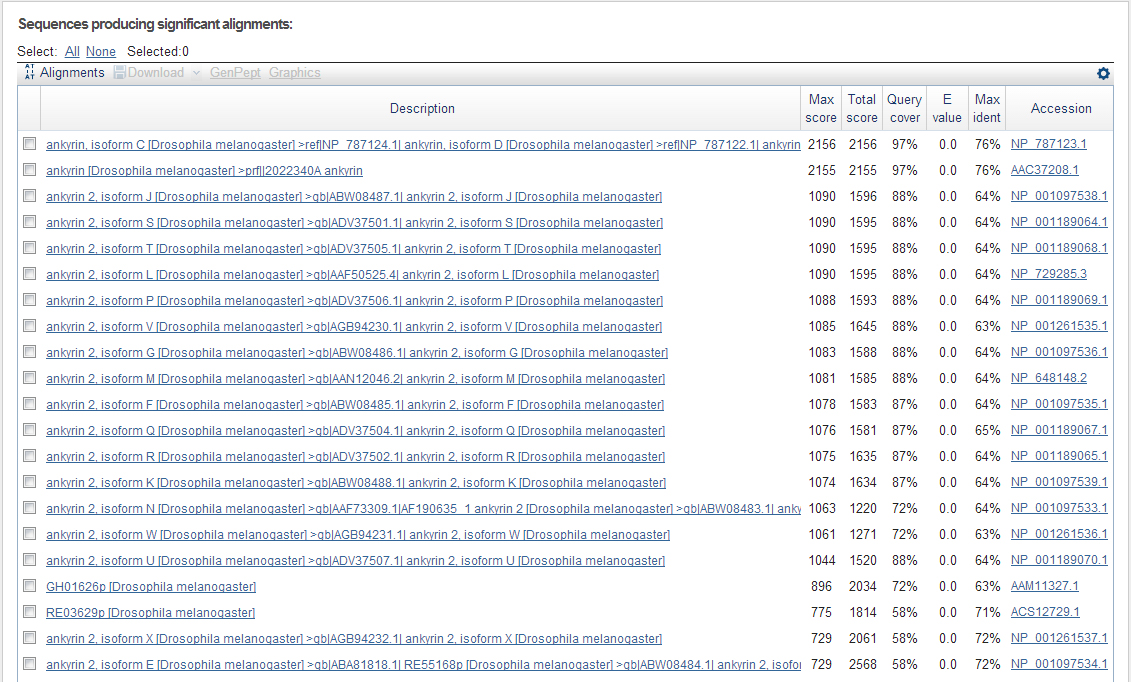
Figure 3: This is my alignment, it seems no matter what I try, I can only get this one alignment with my Contig and Ankyrin. We talked about this briefly in class and you said to not worry about this because we'll be moving onto getting the specific alignments for each exon next.

Figure 4: This is the Flybase GBrowse for Ankyrin and my Contig. I think my contig contains the entire Ankyrin gene and the four isoforms of Ankyrin, Ank PE, PB, PC and PD. I'm unsure if this image represents that the tail end of the gene is off of my contig, but the spliced isoforms are within my contig?
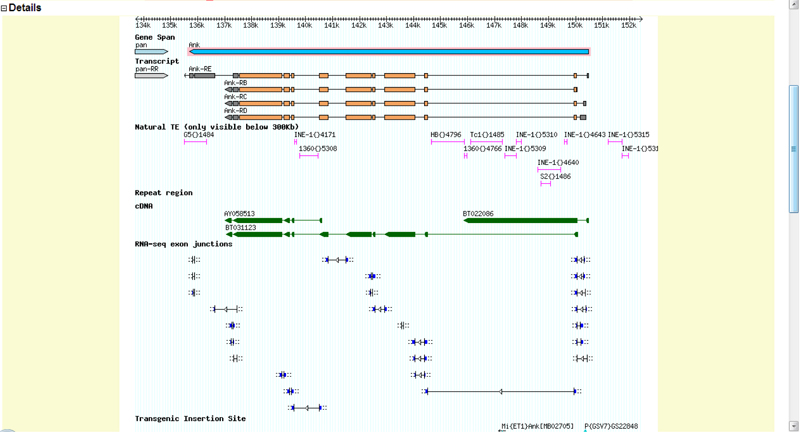
Figure 5: This is the result of my FlyBase BLAST, it shows the graphical alignment of the four isoforms to my contig.
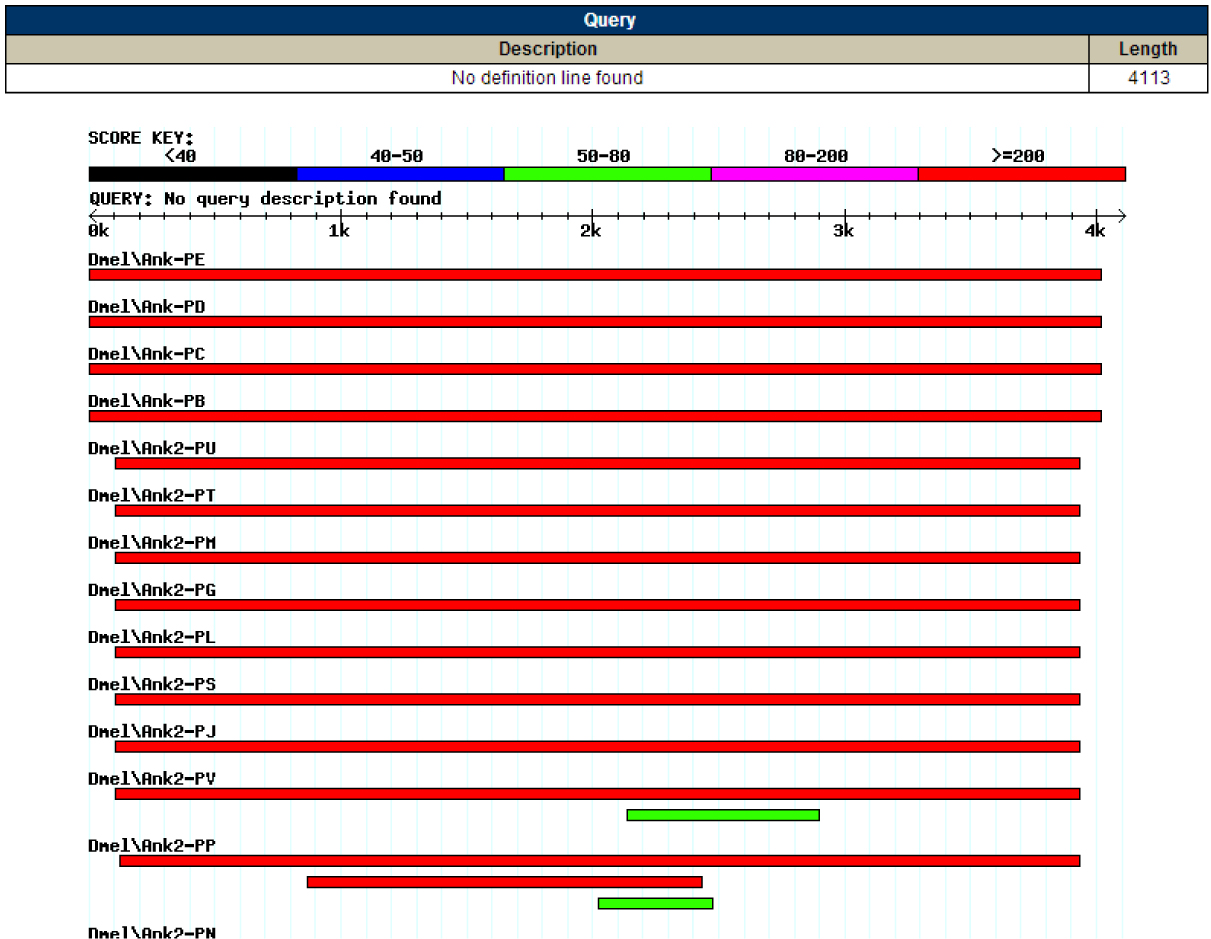
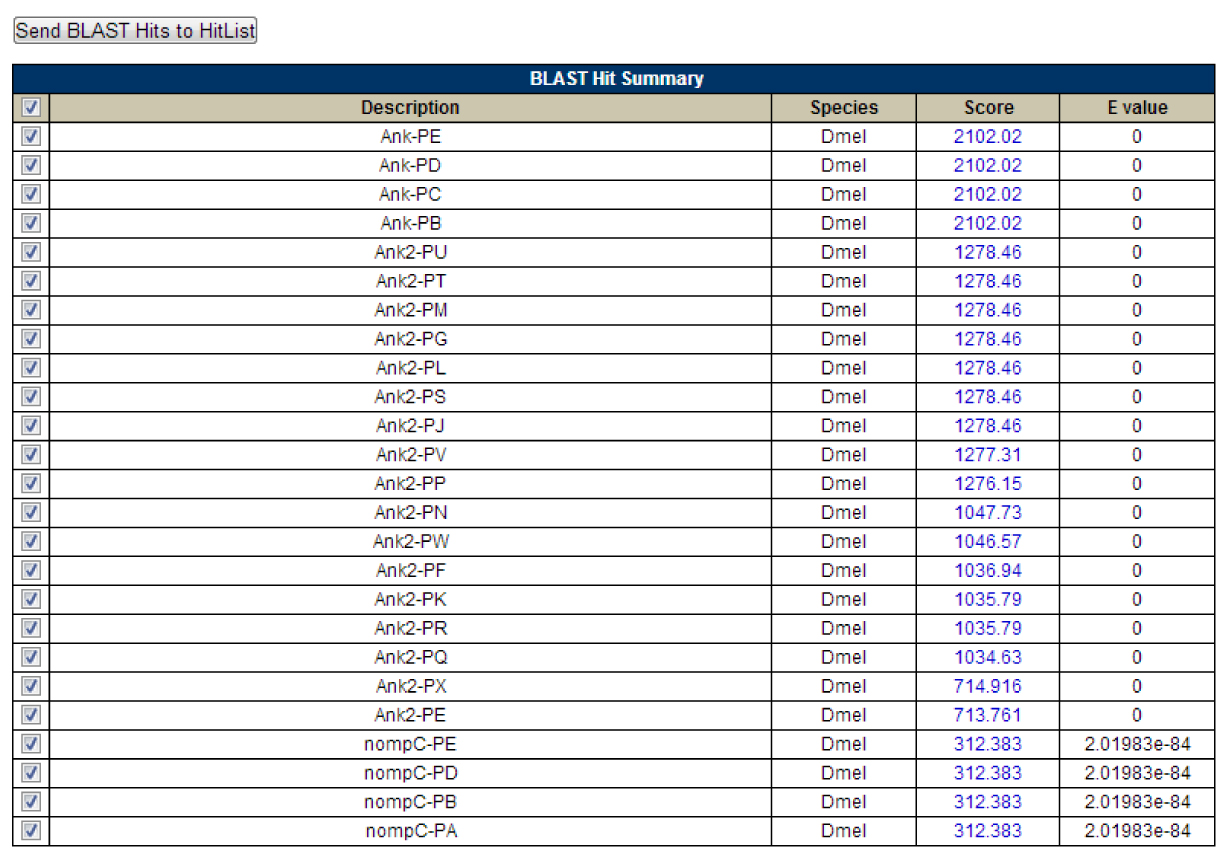
Figure 6: Here is the summary table for my FlyBase Blast, again note the four isoforms of Ank as the best alignments and the Ank2 hits following subsequently.
NOT PICTURED: My GenScan sequence, which I pulled from the analysis folder for my Contig 6, shows a single protein for Contig 6.
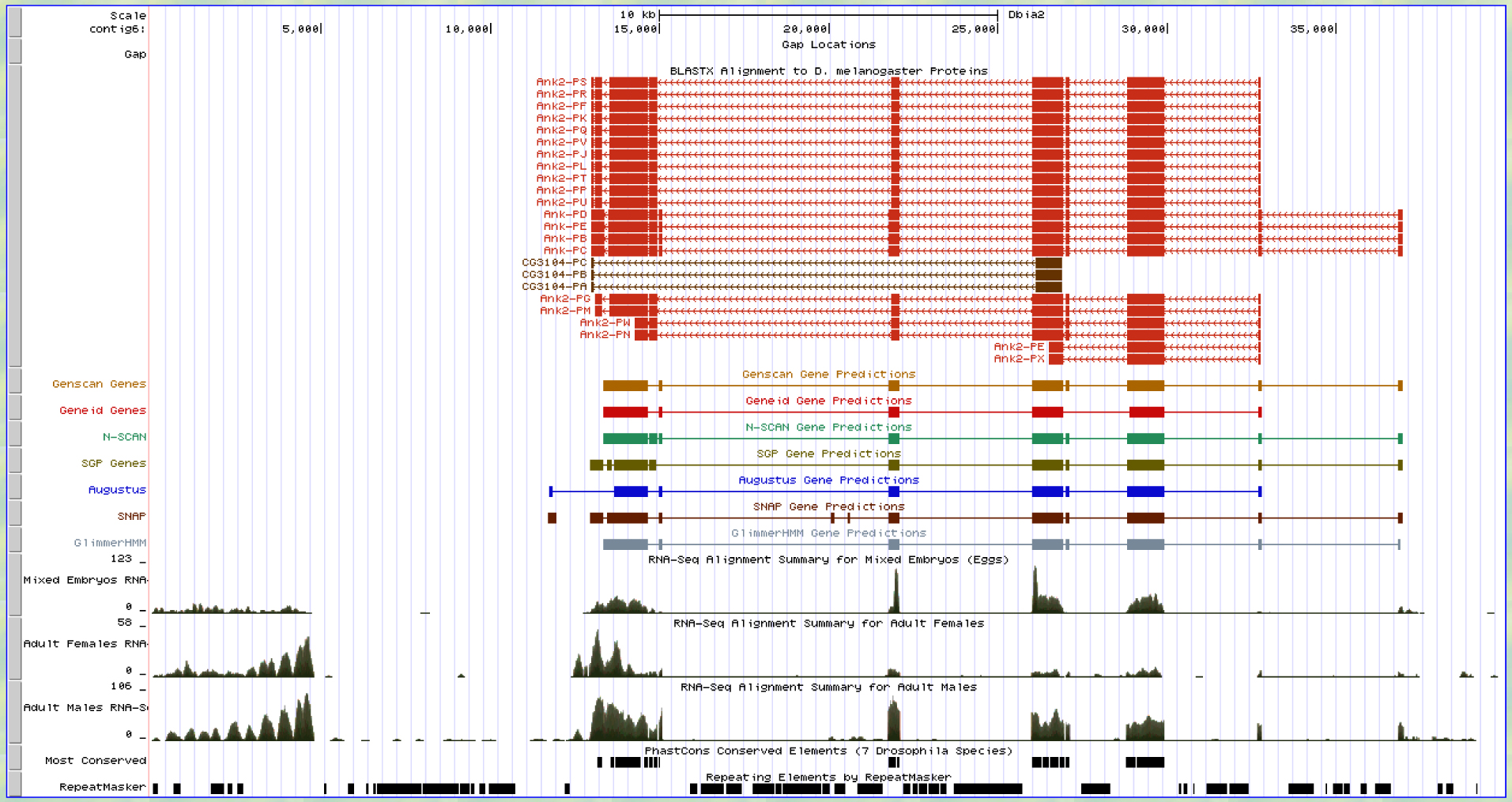
Figure 7: This is a look at my contig in the UCSC Genome Browser at GEP. The BLASTx actually aligns my contig to Ank2 before the Ank isoforms, but you and I worked on this in class and did some extra BLASTs and looked up the locations of Ank and Ank2 to ensure I should stick with Ank. If you go down 10 matches you see the beginning of the alignments for the four isoforms of Ank (PD, PE, PC, PB). The results of the BLASTx show that there are 10 exons for these isoforms. Genscan predicts 8 exons and the other gene predictors agree more or less to the Ank GENE and NOT the Ank2 gene isoforms.