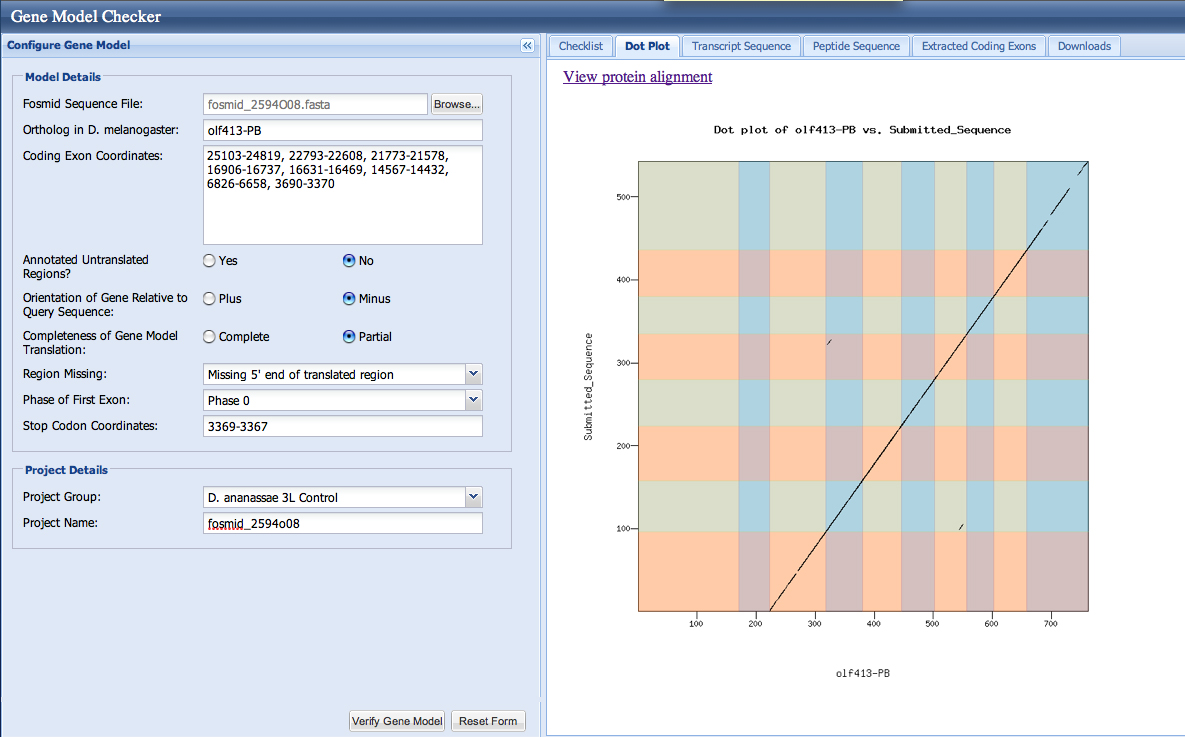
olf413 sequence Gene Record Finder. Gives the 10 exon pieces.
>olf413:2_701_0 MAHPRKAVATPATLQLGPPAQTAQSPAATLRHSRTASSSRRLSLIRCFIS CHTFNLFLLLLLLASGVRAASKLATRSNKTSGSSTASGVGAGTAATSAAA AAASGTPIWDHAIDLNDDFRILWQIINQDITFEIQARTLGYVGFGFSPDG NLAGADMAIGWVDKGQTYFQ >olf413:3_701_0 DRHVTRNGDPEPVVDPSQDYMLMLGYENATHTVLRFRRKLDTCDPSHDIA IT >olf413:4_701_0 NDTMRLLYMYHAQDPPHGSVRPGTLPDPARAFRPYRPMVLMQRAQLPMPS PTHDERVRVLELRNEDVELPAGDLPLFWCKMFKLEDINRKHHLIR >olf413:5_701_0 YEPIYDSSSSVHYLQHITLHECQGAHAELEEMAREQGRPCLGARSIPLAC NAIVASWSRGSE >olf413:6_701_0 GFTYPHEAGYPIESRQAKYYLMETHYNNLKPDFAQLHARQMADNSGLKIY FTHVLRPNDAGTLSI >olf413:7_701_2 MDPNWRHIIPPGQKRVVSEGQCIEDCTGYAFPQQGINIFAVMMRTHQIGK EVKLRQ >olf413:8_701_0 IRQTEELPPIAHDSNIDVAYQDFRRLPQSVHSMPGDRLIAECIYDSSSRK AITL >olf413:9_701_2 GLTMKEESCTVLTLYYPRQKKLTTCHSLPSLPTVLHSLGIEQLA >olf413:10_701_1 DSNPVLISSPPELAGMTLEARLISYDWENQFGEFQEATRKGSFKPICWGA KNHVVP >olf413:12_701_0 GSEFLEGYSINVTKTYKKHRRCKPKRPLAPPTERTAPPPASDLSELPVLH ELDNNNIIEGAARSSRSSATDVHSLSRGSGRHFISCLLWLGASSWWLLLM LRT*
2 of them are low E value and on opposite strand. This could be due to that I do not have the entire gene on my fosmid and these exons are on a different strand that may start the gene somewhere else.
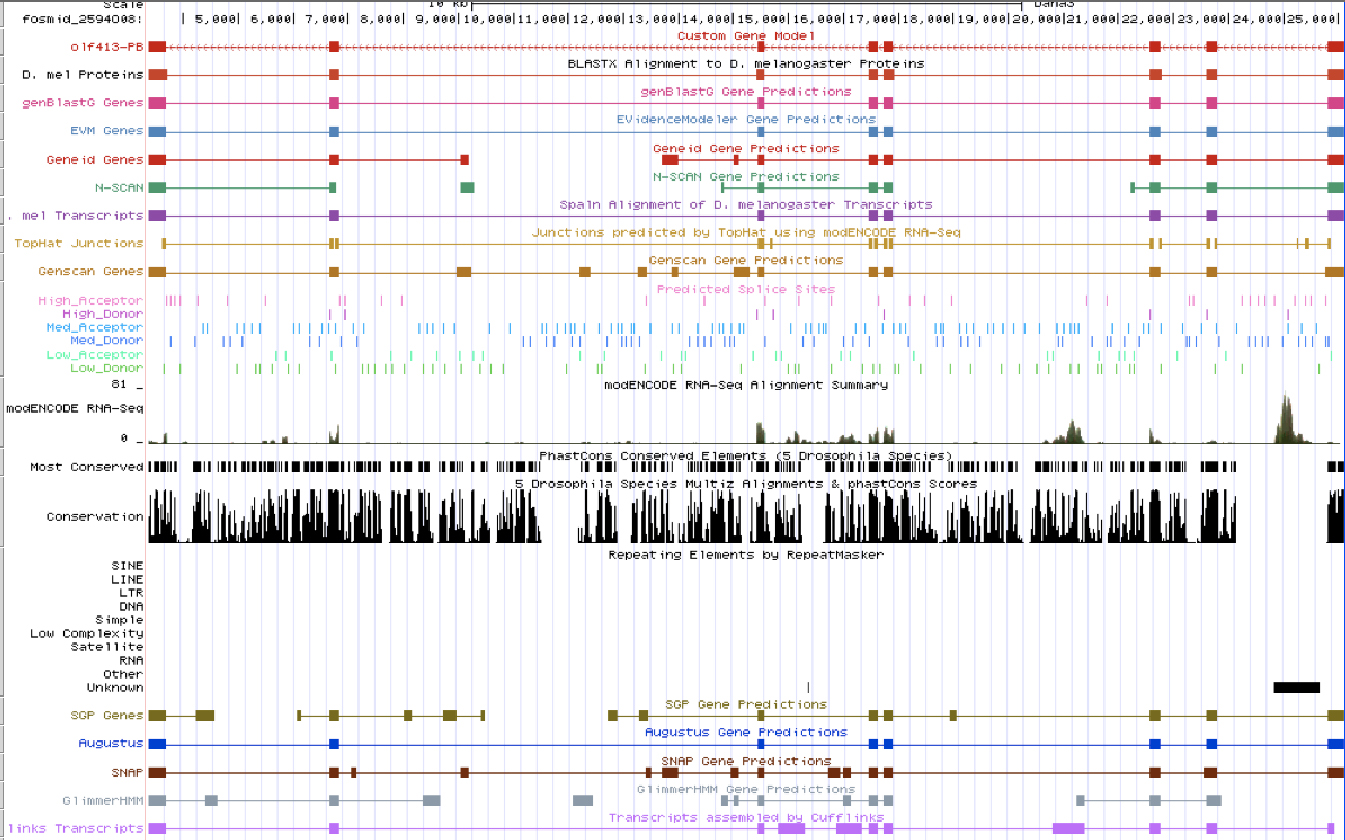
Drive alignment is the table produced by the gene browser and zooming in to the exact positions and finding splice sites and TopHat data to confirm base length and frame alignment.

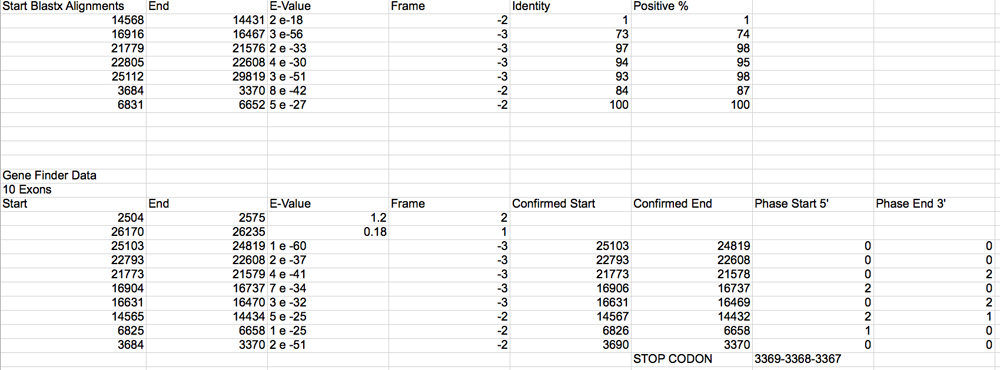
Here is the confirmed data from the Drive table above in the Gene Model Checker. This shows that these exons are confirmed. Also shows that my gene is lacking 5' start to the olf413 gene but ends in my fosmid.
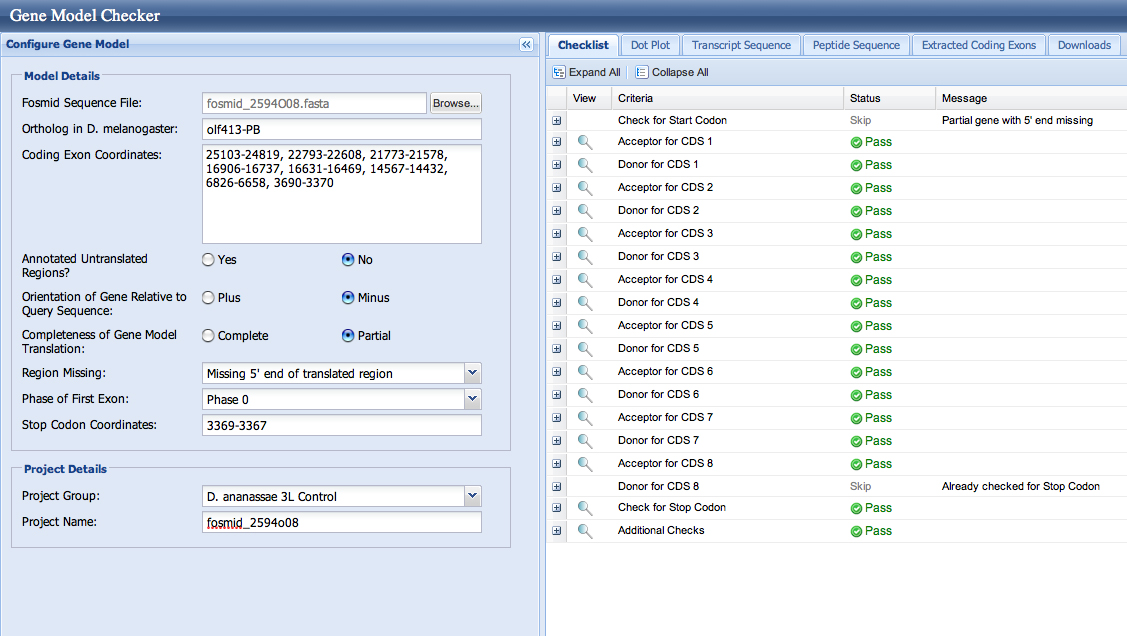
Here is the Dot Plot based on the Model provided in the checker.