| Summary | |
| BLASTX Analysis Graphic summary | |
| BLASTX Analysis Descriptions | |
| BLASTX Analysis Unc-13 alignment | |
| BLASTX Analysis eIF4G alignment | |
| BLASTX Analysis at FlyBase | |
| GENSCAN Analysis | |
| UCSC Genome Browser at GEP |
D. biarmipes contig50 encodes two genes: the D. biarmipes orthologs of the D. melanogaster genes Unc-13 and eIF4G. GENSCAN predicts one peptide encoded on the contig. It has fused the two peptides, making an invalid prediction.
I used the file contig50.fasta from the src folder in the GEP project file for D. biarmipes contig50 as a query sequence in a BLASTX search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings.
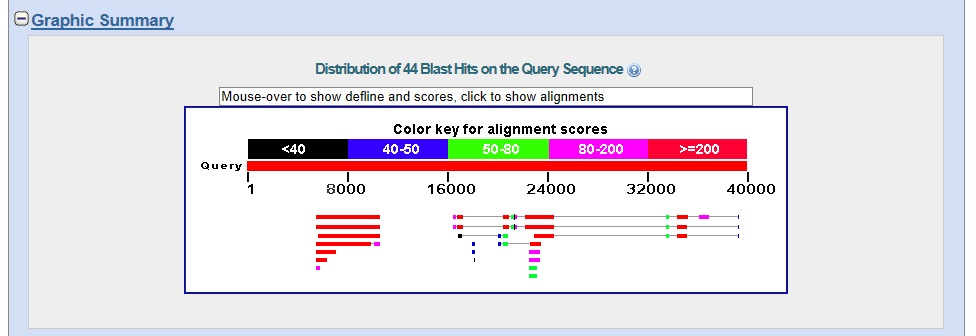
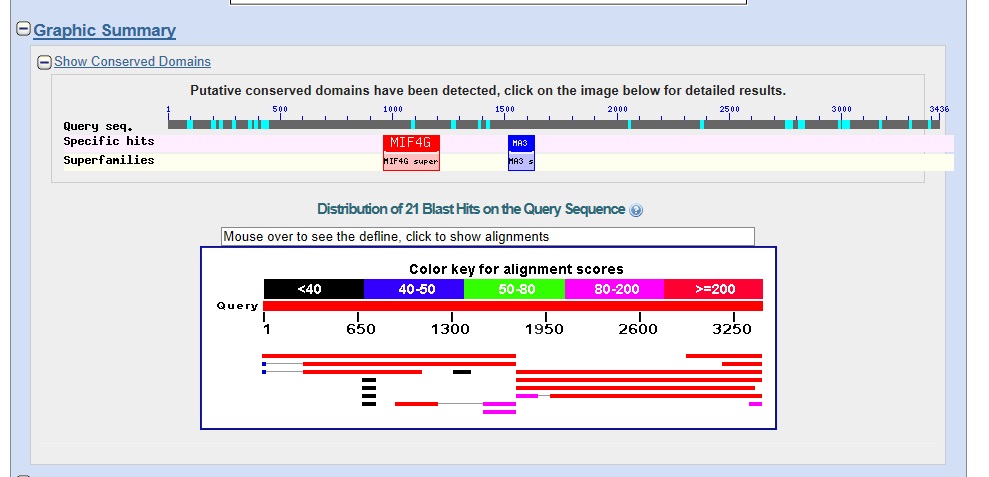
Graphic summary. The graphic summary is shown below.

The results suggest that there are two genes on the contig with protein sequence similarity to the D. melanogaster protein set.
The left gene (Gene A) has multiple good matches to isoforms derived from alternative splicing of D. melanogaster proteins.
The right gene (Gene B) has three good matches to isoforms derived from alternative splicing of D. melanogaster proteins, with five additional hits at lower scores, covering only a portion of the top hit.
Descriptions. All descriptions with E values smaller than e-10 are shown below.
Gene A. The first hit on the description list is Unc-13, isoform C. The coordinates of the alignment are shown in the table below.
| NP_726614.2 | contig | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 3 | 1632 | 10699 | 5636 | -1 | 0.0 | 56% | 67% |
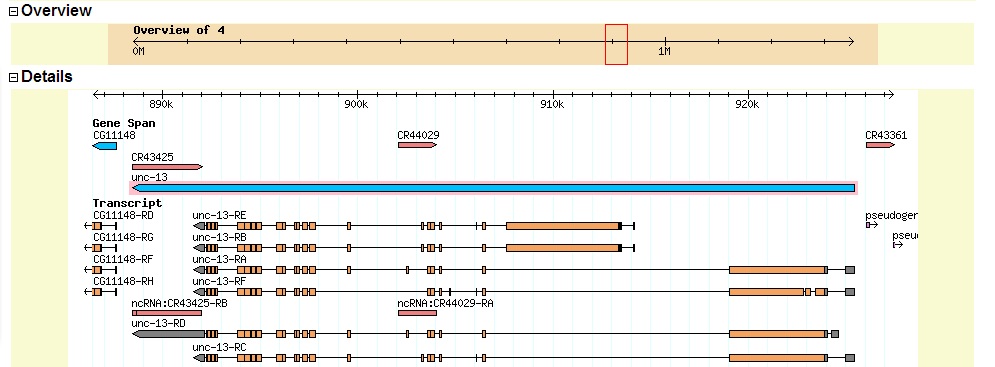
Summary of Gene A: Gene A is the ortholog of the D. melanogaster Unc-13 gene. The D. biarmipes Unc-13 gene is on the minus strand.
Gene B. The coordinates of the top hit (eukaryotic translation initiation factor 4G, isoform B) are shown in the table below.
| NP_001096852.1 | contig | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 611 | 1381 | 24633 | 22318 | -2 | 0.0 | 75% | 83% |
| 1514 | 1675 | 21072 | 20581 | -2 | 2e-74 | 79% | 89% |
| 1741 | 1866 | 17352 | 16915 | -2 | 1e-62 | 79% | 84% |
| 273 | 556 | 35352 | 34441 | -2 | 4e-58 | 75% | 82% |
| 20 | 262 | 37001 | 36261 | -3 | 1e-49 | 63% | 74% |
| 1379 | 1459 | 21699 | 21454 | -2 | 2e-38 | 84% | 91% |
| 1458 | 1511 | 21403 | 21236 | -1 | 2e-38 | 84% | 91% |
| 1866 | 1919 | 16783 | 16562 | -1 | 5e-19 | 70% | 72% |
| 534 | 594 | 33805 | 33623 | -1 | 1e-11 | 66% | 73% |
| 1 | 27 | 39443 | 39363 | -3 | 0.001 | 81% | 88% |
Reordering these in order of the segments of NP_001096852.1 gives the results shown below. Alignments with E values larger than e-10 are grayed out.
| NP_001096852.1 | contig | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 1 | 27 | 39443 | 39363 | -3 | 0.001 | 81% | 88% |
| 20 | 262 | 37001 | 36261 | -3 | 1e-49 | 63% | 74% |
| 273 | 556 | 35352 | 34441 | -2 | 4e-58 | 75% | 82% |
| 534 | 594 | 33805 | 33623 | -1 | 1e-11 | 66% | 73% |
| 611 | 1381 | 24633 | 22318 | -2 | 0.0 | 75% | 83% |
| 1379 | 1459 | 21699 | 21454 | -2 | 2e-38 | 84% | 91% |
| 1458 | 1511 | 21403 | 21236 | -1 | 2e-38 | 84% | 91% |
| 1514 | 1675 | 21072 | 20581 | -2 | 2e-74 | 79% | 89% |
| 1741 | 1866 | 17352 | 16915 | -2 | 1e-62 | 79% | 84% |
| 1866 | 1919 | 16783 | 16562 | -1 | 5e-19 | 70% | 72% |
Summary of Gene B: Gene B appears to be the D. biarmipes ortholog of the D. melanogaster gene eIF4G. The gene is on the minus strand (all matching reading frames are -1, -2, or -3). A view of D. melanogaster eIF4G from FlyBase GBrowse is shown below.
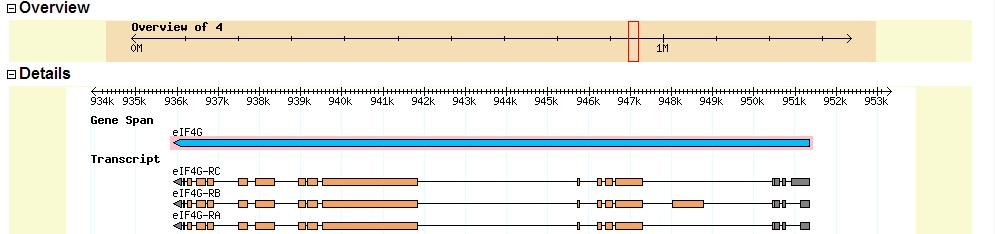
The D. melanogaster eIF4G gene has three isoforms derived from alternative splicing.
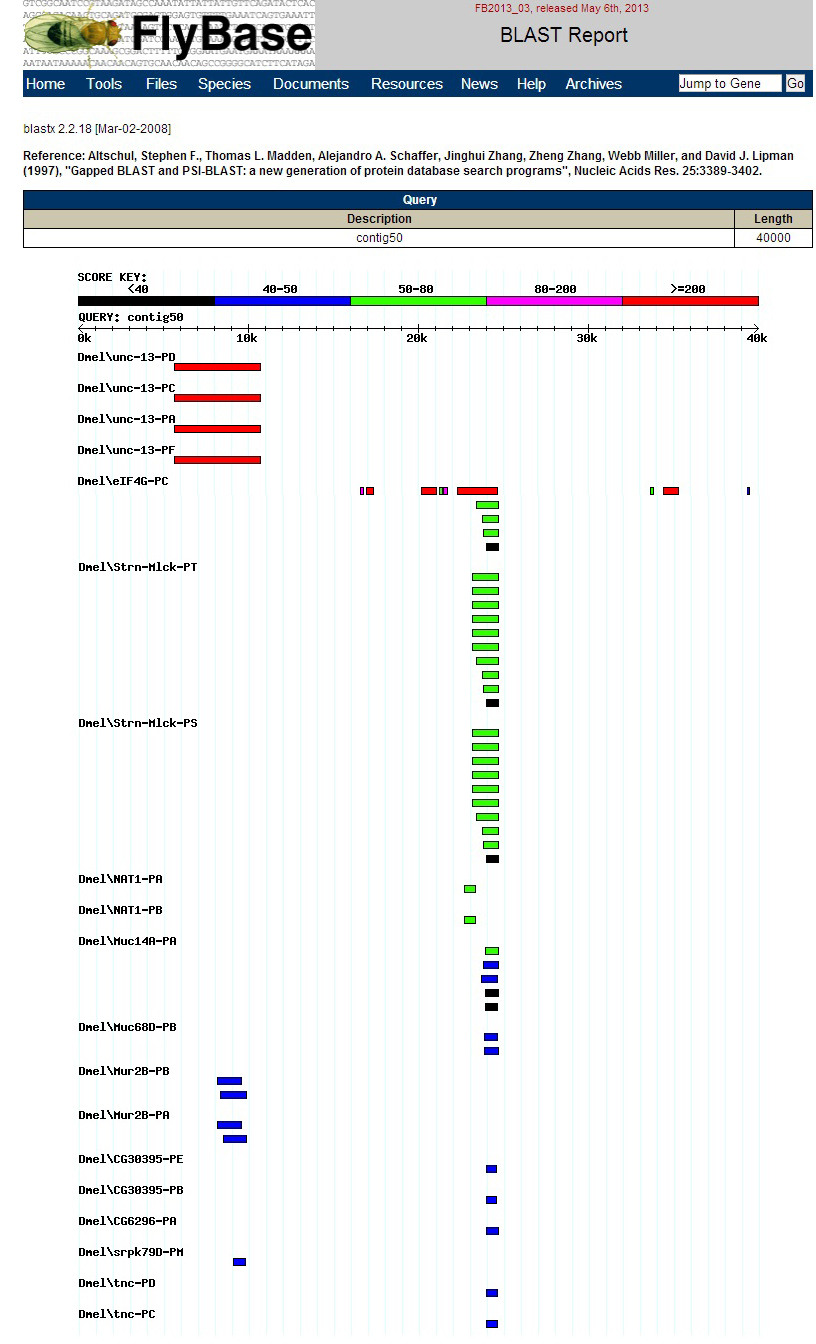
I used FlyBase BLAST to analyze the contig. I set the Database to Annotated proteins (AA), the Program to BLASTX, and uploaded the contig sequence. I restricted the species to D. melanogaster and clicked BLAST.
The graphic output is shown below.
The summary table is also useful, shown below.
The GENSCAN results from analysis/Genefinder/Genscan in the project folder predict one protein on the contig:
>contig50|GENSCAN_predicted_peptide_1|3436_aa MQQAIPTISTQSDIAKIMQPHSAQNMILPANKKTKKYAQQVLPSKPQSLQTMQLQHNHQT PQPQFQINKAYNVVSILKATAQNAQQSSHLTHQQQPSLLLQQTQQHQQSYANVVNRSIPG SGPVGAHQSTVICNGSNIMTVNSCQLNSGDVNSTAIYNLSNQRGLPGSQDGNVRFLNVPD TTKKGNNLGASVVSSNSTTGVGNGTTSCTGVGITNNSQITLTSSHIGTTMGVALGTTAAG TTYMHEKNIVGVSVSCVDTSRKYDFKNSSLIKNNSFQAAAEYVSTGNNSSGNSRSNPQSG AIFRGPPPTANTPRGATSGATRHVHVQPMYSQPLHQNMVIQQYTQYNPRQQTFPTSHLQY APAPMPYYQYQYVPTIQQQPPPHTRSAVSVSTNVNVGNTLQPVQSGPNGPLTAPGSTSSQ LQLITSTVQPGTNNVMGVGGPGSGMGQTNSNTDNDFSEQVTLPNTPTVVLSEGQIRIPQQ DTVGINNLSNTTSQGSETRTNASYTPVEPIPISRQDVGQTPIVSAMSDAPSVEILPTPQR GRSKKIPIVSPKNASEASAAPTTDETDDALSKPIVTTAKAPTEQSLAHQKLLTSESPQQK QSVSNTEITKDEPTKLEDIKIDELDSVVSSGNLQTELLSFNVKDSQPPSNFSEEPETAST VEIPPLDFIEDSSKMHTALDNSESTLSIEILEKSTVESFKDNQSAEQQTQQDINLRSVPD ETEISSMALKEVTTLDNRQTENKDTIKSKNNADISKELTRETTMDSLLKNNTDEVVEHQS GTSTDSKPEEDLEDRLQSTDQKLEGTGITVSSFINYNEGQWSPSNPTGKKQYNREQLLQL REVKASRKQPEVKNISILPQPNLMPSFIRNNNNKRVQSMVGIIGNRSSESGGNYIGKQIS MSGVQGGGGRSSMKGMIHVNLSLNQDVKLSENENAWRPRGLNKSDGDSEAKSTHEKDELI RRVRGILNKLTPERFDTLVEEIIKLKIDTPEKMDEVIVLVFEKAIDEPNFSVSYARLCHR LISEVKGRDERMESGTKSNLAHFRNALLDKTEREFTQNVSQSTAKEKKLQPIVDKIKKCK DANEKAELEAFLEEEERKIRRRSGGTVRFIGELFKISMLTGKIIYSCIDTLLNPHSEDML ECLCKLLTTVGAKFEQTPVNSKDPSRCYSLEKSITKMQAIASKTDKDGAKVSSRVRFMLQ DVIDLRKNKWQTSRNEAPKTMGQIEKEAKNEQLSAQYFGTLSSTTPVGSQGGSGKRDDRG NTRYGDSRSGSGYGGSHSQRSDNGNLRHQQQNNTGGAGHSNGNNDDNTWHVQTSKGSRSQ AVDSNKLEGLSKLSDQNLETKKMGGLGQFLWPSNTTRQSSASTSTPSNPFAVLSSLIDKN GSDRDRDRDRSGPRNKGSYNKGSIERDRFDRGIHSRTGSSQGSRENSSSRAGQHGQGRSL LSSTVQKSTSHSKYTQQAPPTRHAGKTPTSLVSSNVNTGGLYRGSEQQSPTSATFSQGSR SVAPVAVFKEAGETELKLIKSVVSEMIELAAASKAVTPGVVSCMNRVPEDLRCSFLYYLL TDYLHLANVGKQYRRYLAIAVFQLIQQNYISVDHFRLAYNEFSEYANDLIVDIPELWLYI LQFAGPLIVKKILTLSDVWNKNLKDNSPSSVAKKFLKTYLIYCTQDVGPKFARSMWSKFN LKWSDFMPESEVSDFIKSNRLEYIENESKSPVIEQRESPEKHVKNVIDHIEHLLKEGTTA DCIIDYSNHYARHDYFHNTQNGALSSDTGRSPYSHIPYRAQNSREYYAEPYDLGNHGLEE YSSECHLTSDRVLTTIDKRNNSYEYDYIECYEAQEQRDVESESIDNWNENHSGVGAQYGF EYANQKCTSAKVLPTLPVNKTGSGPSKPSATQMDIIFKTKGMCIEKDQRFGVCMAKADEY DLRILPGDYQNIYADNLNGYAGFAYPSTFLNNAVPAAPSRALPQTNRSSFYLGQDIFGLN ADEAQREEQSKCDFGMDQAVTMDSGSTSYDVLEKMSRPYTSMLPLDYSDYHDSYYNTDNL STYSDTPPTTNSQLKLQKQRKLSLMMAMTTASVIASGETRVAVHSKHSKKPTEFQTDSIL GNNISTNAATKASDRFLETECSGGIVTLSGPGAVTGTPPVALSTIIKTRKLPKVLPAPQF NSSLHLNSSTANALSSPVYSSDTAAEKSHRPKQLPKLPTSLPQIKPYSIHNSNLTTLSAA DELPSYSLKSNTASSPLAITETVATTSYLSSTETKTCPSKKPNEVYLTKSIDDGLTPPWT PSPPTQLKQYFSPSAELPSQIVQKNSPTSHLPDIEKAKSDIKPDLHENLNESILECKKSP EPCSEPESALFNISEYPKPYTLDIIPFSGEKENHITNAASTSTTTHSYDDNGELCTELIK LQYSEPSGDSHLFPYFNTWTTSGGNYLPFEVGQGANISTQSYTVTTKAESVMSPLTSFIP PLFISSNFNSKNLTEDIVFSTTSSPHDKNTTVSYSSSNNAFTKTVYVNNPEEVPVTTSEN AIPPSSDLTLRSPSSLVPILSYTDYMKQFELPDLPQPIMELSENIPVTHSDSNSVPINDI ATNAEFSCPPNELDVTSKCSDPLPSYLGELFDAYNVPSLSIENQESPIKGQIGDLFNIAI VPQVPINTIECSVDPPNRFNLLPEADAVENHAVAFDDNFYDSFNVDIKELTASVANIEPE NGSRNFPSSSVENSTDIDDKPSDEFINEKTVGTRDLNQNLSSGQGGYYKPSQAQQKASVV ASAATSVLGGISKGLKGGLDGVFSGVSSTVEVSQTTNSTKKGFSFNLASKLVPSVGGLLS SSSSNSTKQTQGQQMDPTPTFITTSAENFSSESSTANVTMTSPPRLYKNAEDTLYAATVS NITDVNHFYSEGLIDSNSALEGNISDTYDESYNEMILTDKLVDEQIVGRDSGYGLTENPY SYHVSCVGQDDTTKACSQLHDIPIDTSFVIEQAEIKSGLVHFPESTTKKGGTTSGGMFGS ILGKAAAAVQSATHAVNQGASSVASVVSQKQSVVPTAHNIRDLSPNAIKRDSNRDSVGFN VMNVDYSYQLGNEESLSSHYENTGDDYENSNIKMHEYGTYTDNKAFVNYHSNGNQSQFQD DTAIFGQSKVIGNGTKILPTVPPAGSTGKKLPTVNGKSGLLIKQMPTEIYDDESELDELD GSPLIGKKPSYHIDSEQDDYYLGLQQTTPSNQANGYYEHVNNGYDYREDYFNEEDEYKYL EQQREQEKLHQPKNKKYLKQTKGVLSTTQPQSSLDFIDEGQDDDFMYENYHSEEDSGNYL DESSSGSVGPSEGRNLKMDSNGDASLASTSNQMKRDSFTNNSLHKLDTVVGESTSNLTGI IKEKMCSDLDERSEDINDQLSDLTDINKLTLLKKKSLLRGETEEVVGGHMQMIRQPEITA RQRWHWAYNKIIMQLN
The protein was used as a query sequence of a BLASTP search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings. The results can be seen in the graphic below, showing the fusion of two genes by GENSCAN.
Summary of GENSCAN analysis: GENSCAN made an invalid prediction by fusing two genes found by the BLASTX search (Unc-13 and eIF4G).
Here is a view of contig50 in the UCSC Genome Browser at GEP.
BLASTX Alignment of D. melanogaster proteins. The BLASTX track at the top of the image shows alignments to two distinct regions, as was seen in the prior BLASTX analaysis. The leftmost D. biarmipes gene, Unc-13, aligns to four D. melanogaster isoforms produced by the D. melanogaster Unc-13 gene. The rightmost D. biarmipes gene, eIF4G, aligns to three D. melanogaster isoforms produced by the D. melanogaster eIF4G gene. These are the same results seen when the contig is used as a query sequence in a BLASTX search of D. melanogaster proteins. There are two genes on the contig: the D. biarmipes orthologs of Unc-13 and eIF4G.
GENSCAN predictions. Only one GENSCAN is made, as it fuses the two genes.
modENCODE RNA-Seq. Transcripts aligning to Unc-13 and eIF4G are seen.
Conservation. The exons of Unc-13 and eIF4G are clearly conserved. The intergenic regions between Unc-13 and eIF4G are not conserved. The introns in the two genes also do not appear to be conserved.
