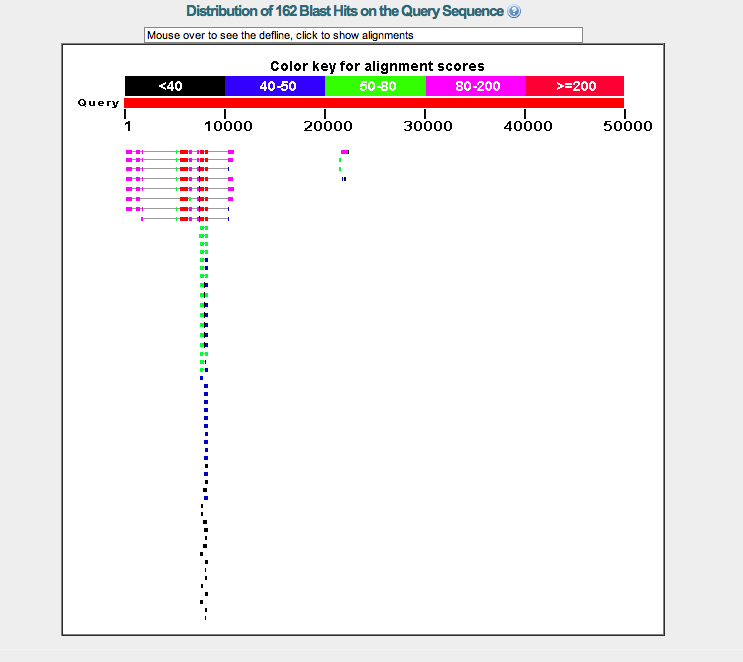
My file [contig29.fasta] containing my Drosophila biarmipes contig was used in a blastx search of nrp's against D. melanogaster's translated nucleotide database at the National Library of Medicine.

The visual alignment map suggested that one gene [Asator] was present in many isoforms on this particular stretch of the D. biarmipes DNA sequence.
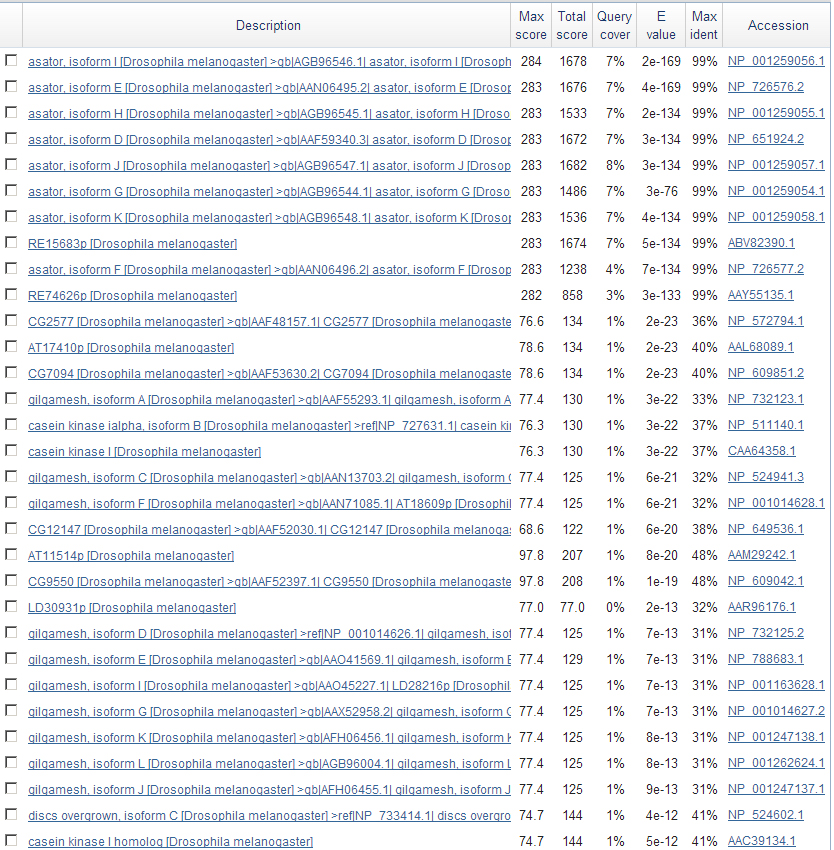
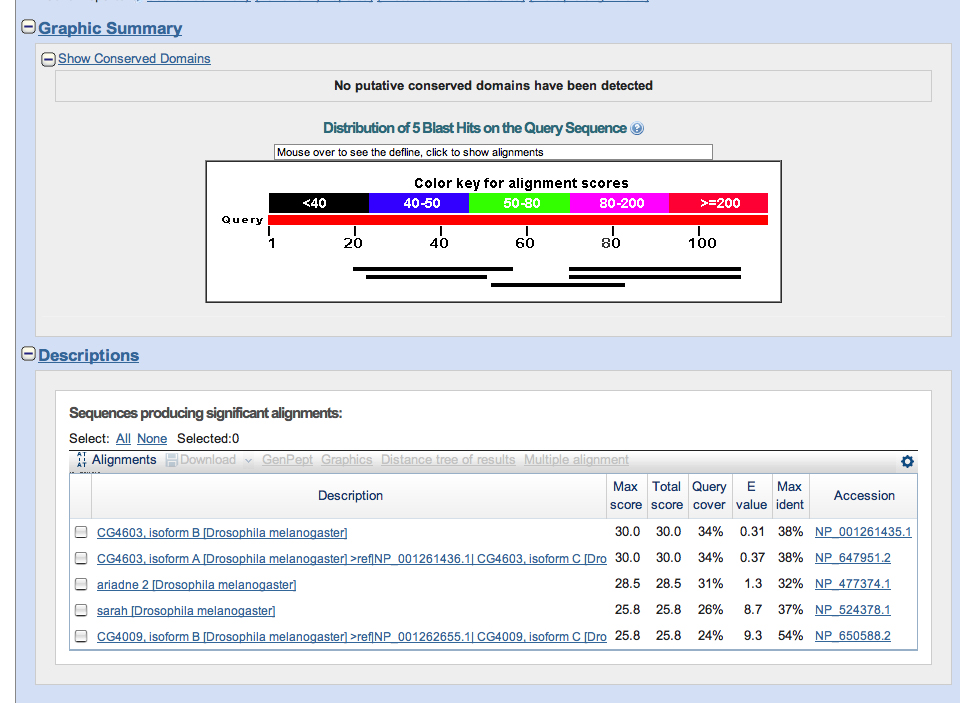
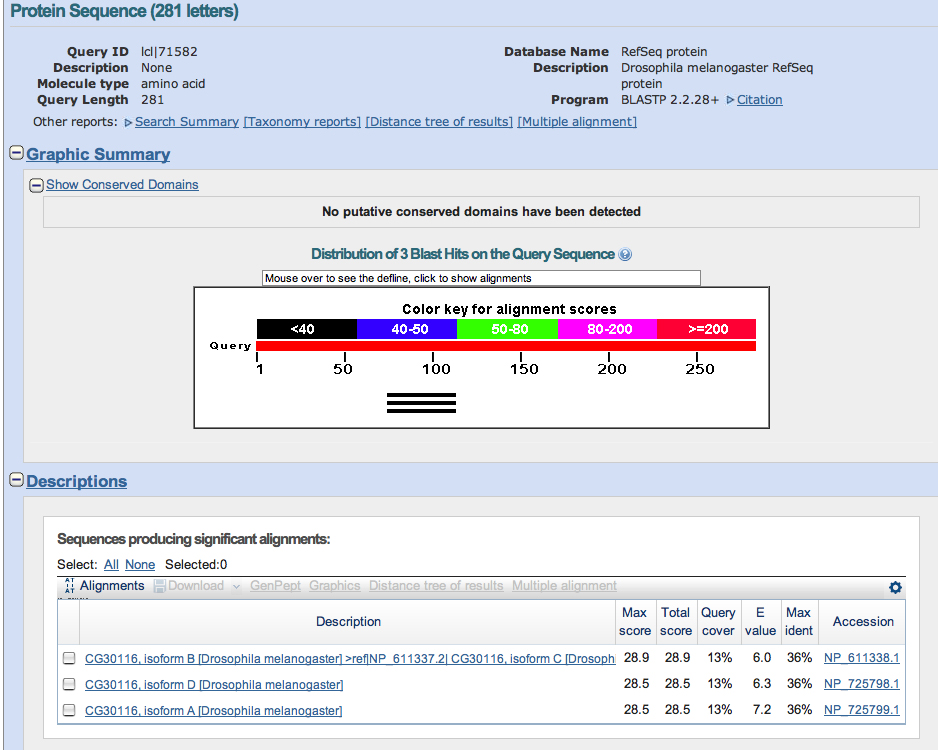
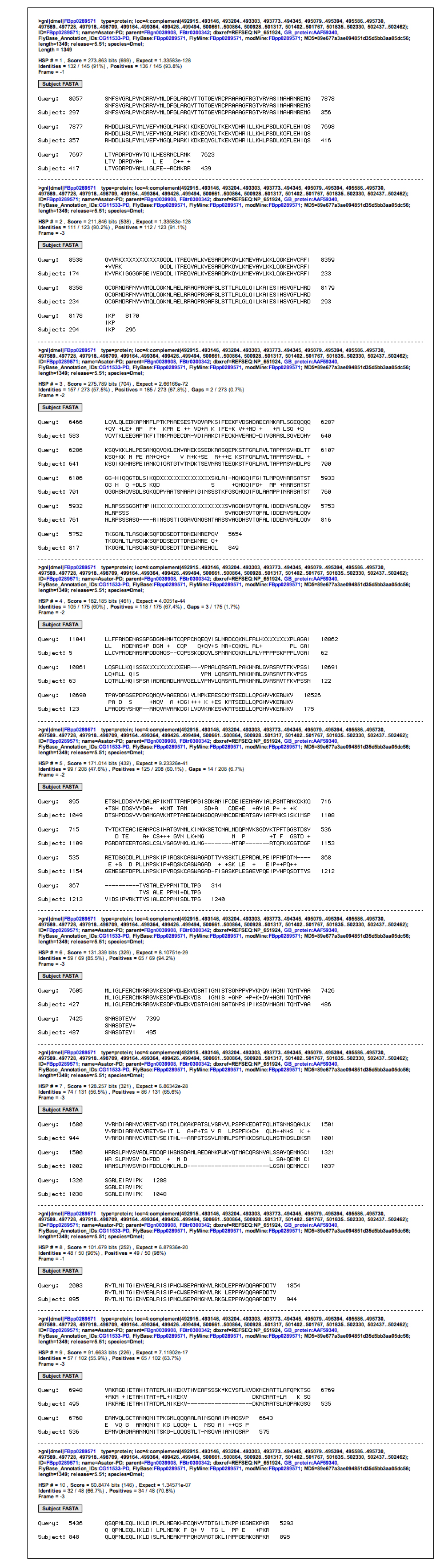
A list of significant findings displayed a few results that may have been significant (Mainly Casein Kinase I and Gilgamesh) but they span the same region as the definite Asator hits.
A useful table -- Primary Asator Hit
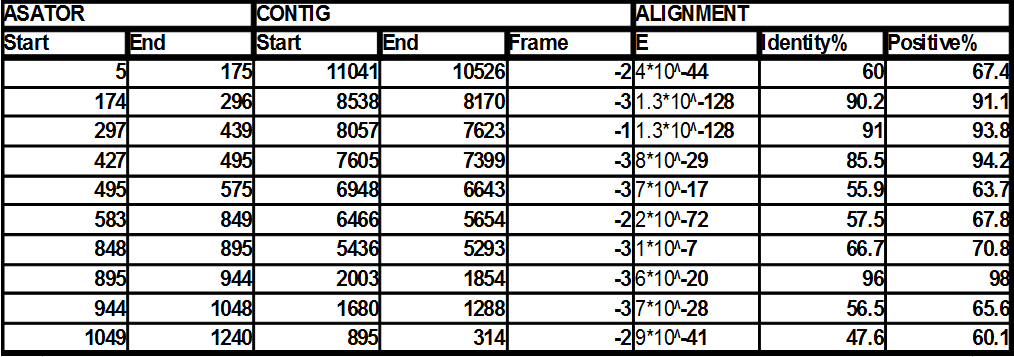
Comparisons of the table and the visual alignments reveal that a section [895-944] is insignificant yet it gets flagged by every isoform except one. Further investigation may reveal more information about this specific segment of DNA.
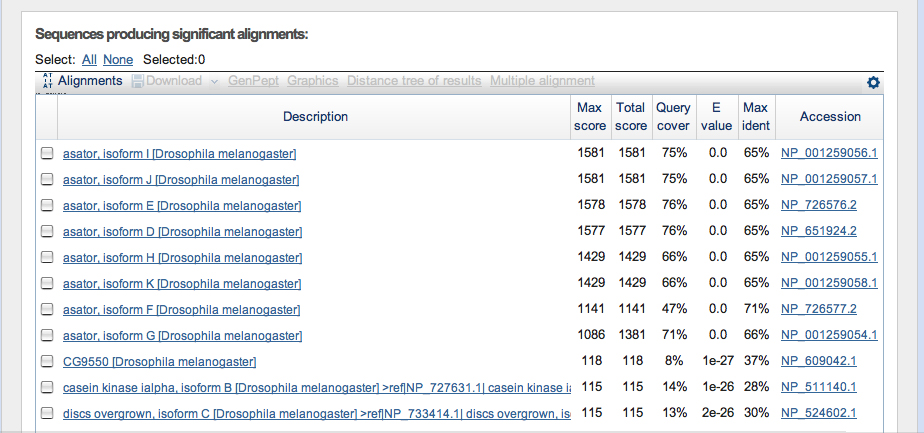
Genscan predicted Asator, Casein Kinase I, and a junk model sequence. Other predicted sequences were followed until they provided junk blastP's.
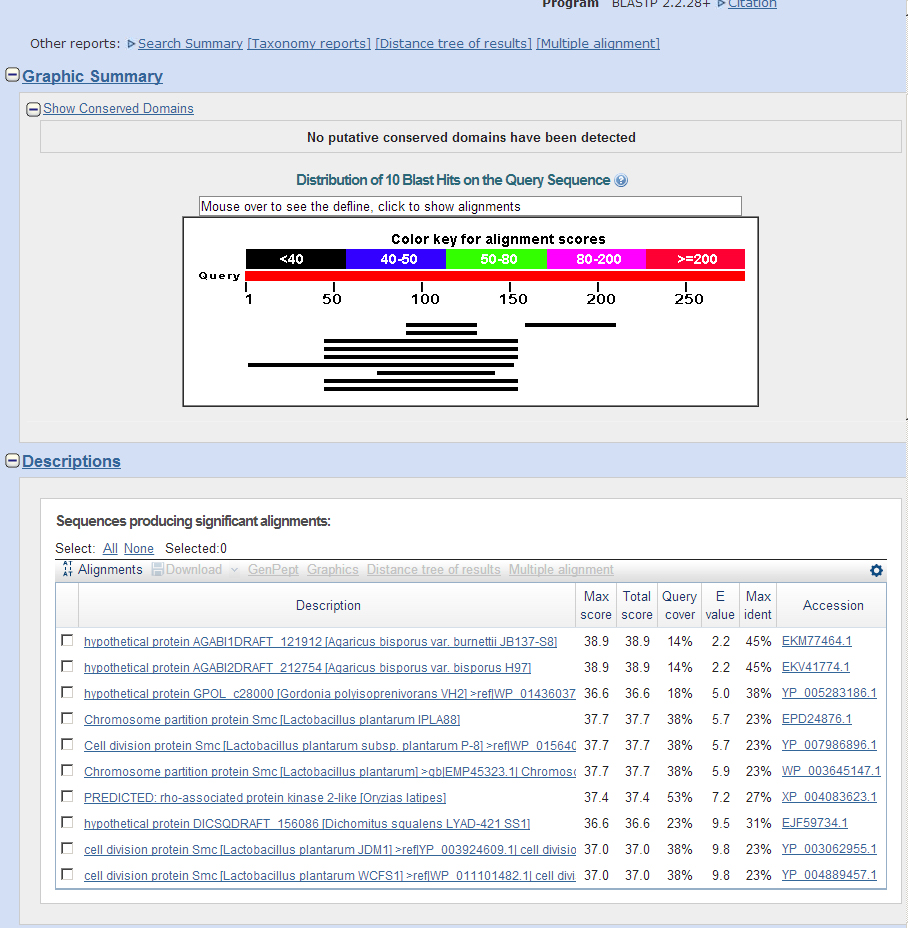
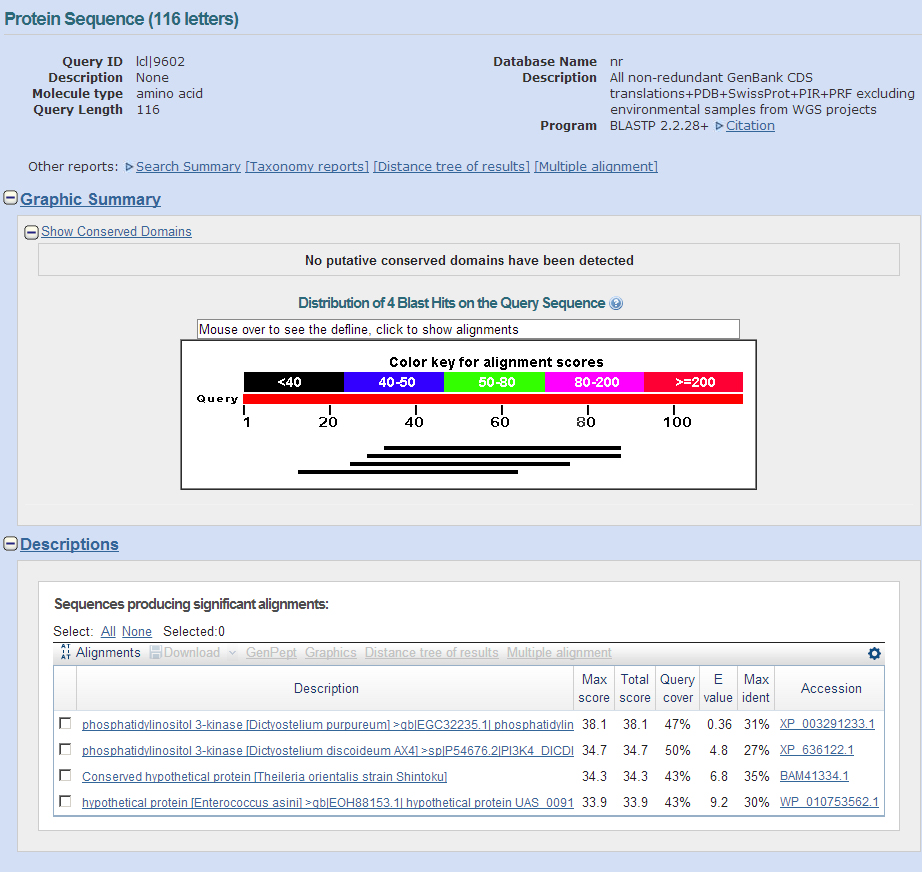
It appears that this contig only contains the Asator gene and its many isoforms.
---alignments for info only [Iso-PD] ----
LIST OF SPECIAL CIRCUMSTANCE --
11_0-3' -- GAIN OF SEQUENCE See: 11_0_META_-3_ALL--NOTE_RNASEQ
11_0-5' -- CUT TO COMMON 5' See: 11_0_5'_ECALL_-3_ALL--NOTE-G
8_0&7-0-3' -- CUT TO COMMON 3' AND MERGE EXONS See: 11_0_3'_ECALL_-3_ALL--EVIDENCE__RNASEQ+HIGHACCEPT
X_ISOPF -- RNASEQ DATA REJECT EXON See: X_FULL_ISOPF-END--NOTE_RNASEQ_REJECT
5_2,6_0_FULL_NESTED_-2_KEJGHDI.JPG
6_0,7_1_FULL_-3_KEJGHDI--NOTE_RNASEQ_MERGE.JPG
8&7_1_FULL_NESTED(5')_-1_KEJHDIF.JPG
9_0_FULL_ECALL_-3_KEJHDIF.JPG
10_0_3'_ECALL_-2_ALL.JPG
10_0_5'_ECALL_-2_ALL.JPG
10_0_FULL_-2_ALL.JPG
11_0_3'_ECALL_-3_ALL--EVIDENCE__RNASEQ+HIGHACCEPT.JPG
11_0_5'_ECALL_-3_ALL--NOTE-G.JPG
11_0_FULL_-3_ALL.JPG
11_0_META_-3_ALL--NOTE_RNASEQ.JPG
12_0_FULL_ECALL_-3_KEJHDIF.JPG
13_0_3'_ECALL_-3_ALL.JPG
13_0_5'_ECALL_-3_ALL.JPG
13_0_FULL_-3_ALL.JPG
14_0_-1,13_0_-3_RNASEQ_REJECT.JPG
14_0_3'_ECALL_-1_ALL.JPG
14_0_5'_ECALL_-1_ALL.JPG
14_0_FULL_-1_ALL.JPG
15_0_3'_ECALL_-3_ALL.JPG
15_0_5'_ECALL_-3_ALL.JPG
15_0_FULL_-3_ALL.JPG
15,14,13 - INTEREST.JPG
16_1_3'_ECALL_-2_EJGDI,16_0_FULL(NESTED3')_ECALL_-2_KHF--START.JPG
16_1_5'_EVIDENCE_-2_EJGDI.JPG
16_1_FULL_-2_EJGDI.JPG
17_0_FULL_ECALL_-2_EGD--START.JPG
19_0_FULL_ECALL_-1_JI--START.JPG
X_FULL_ISOPF-END--NOTE_RNASEQ_REJECT.JPG