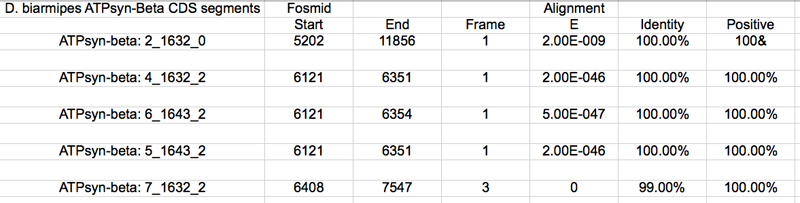
>ATPsyn-beta:2_1632_0 MFALRAASKADKNLLPFLGQL >ATPsyn-beta:4_1632_2 TAPFSARSHAAKAAKAAAAANGKIVAVIGAVVDVQFDDNLPPILNALEVD NRSPRLVLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGYPIRIPVGAE TLGRIINVI >ATPsyn-beta:6_1643_2 RSHAAKAAKAAAAANGKIVAVIGAVVDVQFDDNLPPILNALEVDNRSPRL VLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGYPIRIPVGAETLGRII NVIGK* >ATPsyn-beta:5_1643_2 RSHAAKAAKAAAAANGKIVAVIGAVVDVQFDDNLPPILNALEVDNRSPRL VLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGYPIRIPVGAETLGRII NVI >ATPsyn-beta:7_1632_2 EPIDERGPIDTDKTAAIHAEAPEFVQMSVEQEILVTGIKVVDLLAPYAKG GKIGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYNE MIEGGVISLKDKTSKVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQ DVLLFIDNIFRFTQAGSEVSALLGRIPSAVGYQPTLATDMGSMQERITTT KKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRAIAELGIYPAVD PLDSTSRIMDPNIIGQEHYNVARGVQKILQDYKSLQDIIAILGMDELSEE DKLTVARARKIQRFLSQPFQVAEVFTGHAGKLVPLEQTIKGFSAILAGDY DHLPEVAFYMVGPIEEVVEKADRLAKEAA*
I BLASTx and aligned two or more sequences the query was my contig58.fasta and the subject was each of the above sequences. The results below were selected based on their E-value and genomic location. D. biarmipes ATPsyn-Beta CDS segments

The UCSC genome browser below displays the 5' end of D. biarmipes ATPsyn-Beta. The ATG codon (MET) spans 5202-5204 in reading frame +1 which was listed in the BLASTx results.
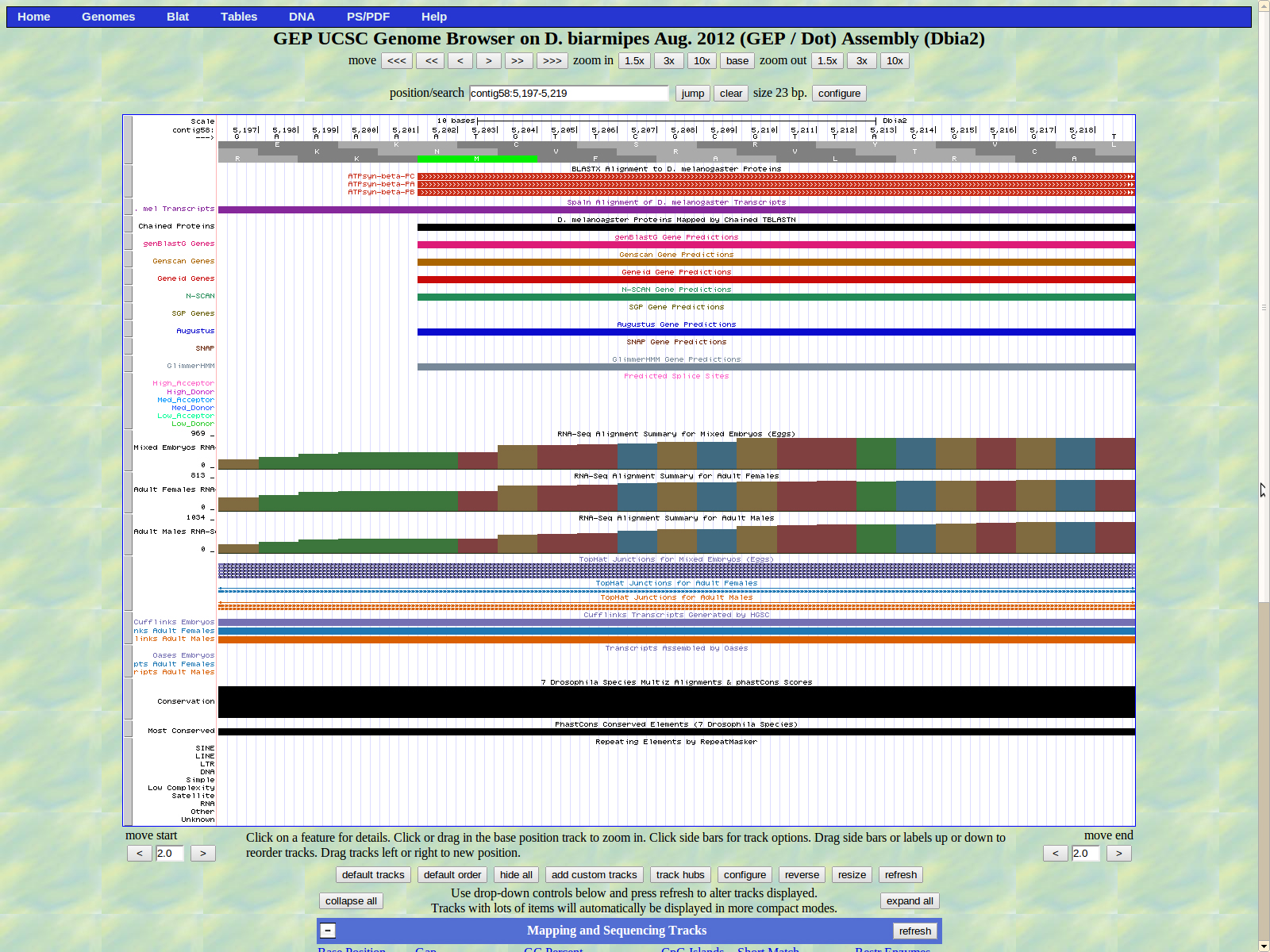
The first of two introns is displayed below. The last base of the first exon (left) is 5265. We have a high donor GT splice site is located at 5266-5267.
The first base of the second exon (right) is 6041. The high acceptor AG splice site is located 6039-6040.
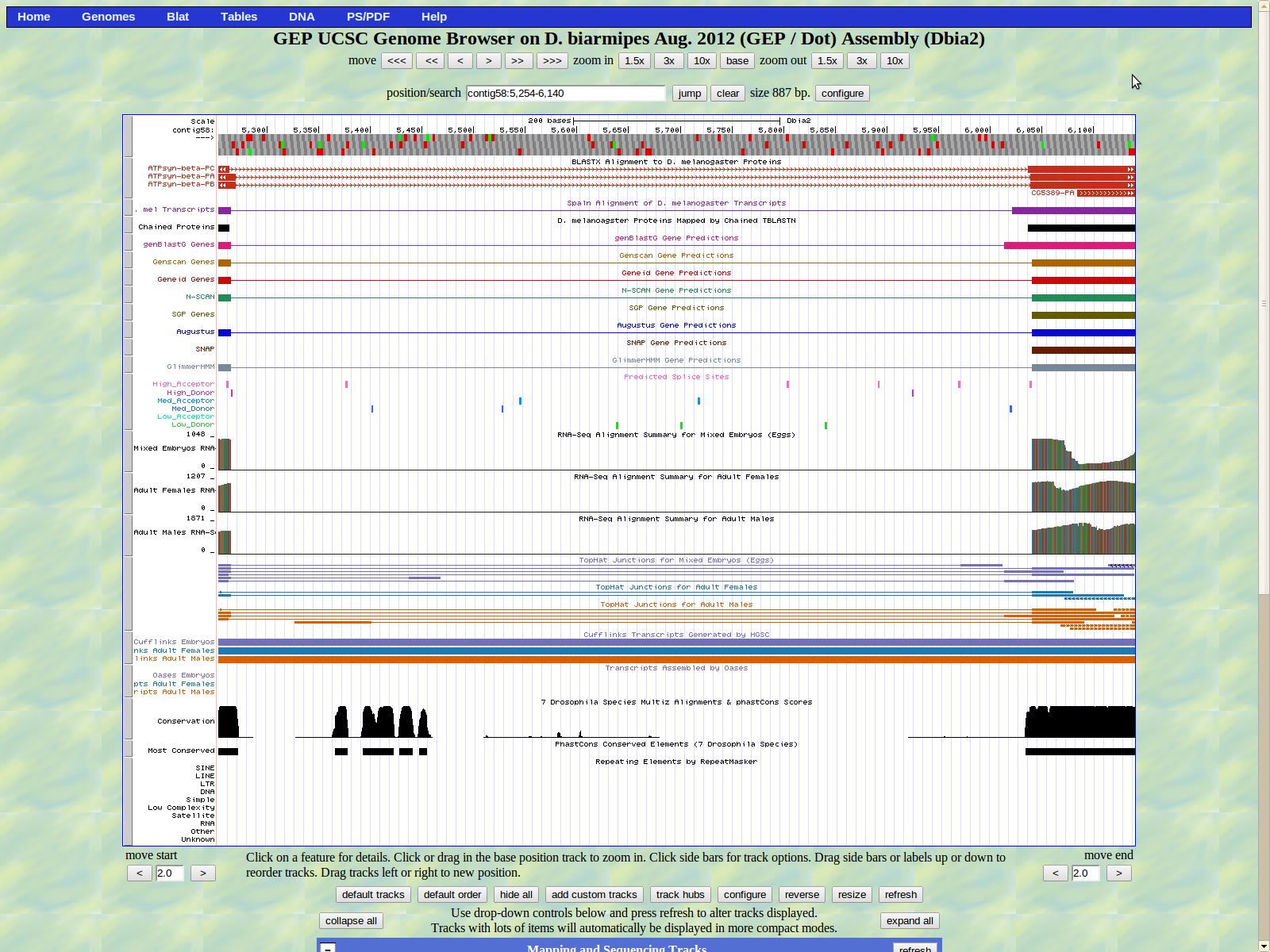
Shown below is the second intron of ATPsyn-Beta. The last base of the second exon (right) is 6352. This time a medium donor GT splice site is shown at 6353-6354. Also this is the final exon of isoform PB.
The two remaining isoforms PA and PC display the last exon (left) beginning at 6406. The high acceptor splice site AG is located at 6404-6405.
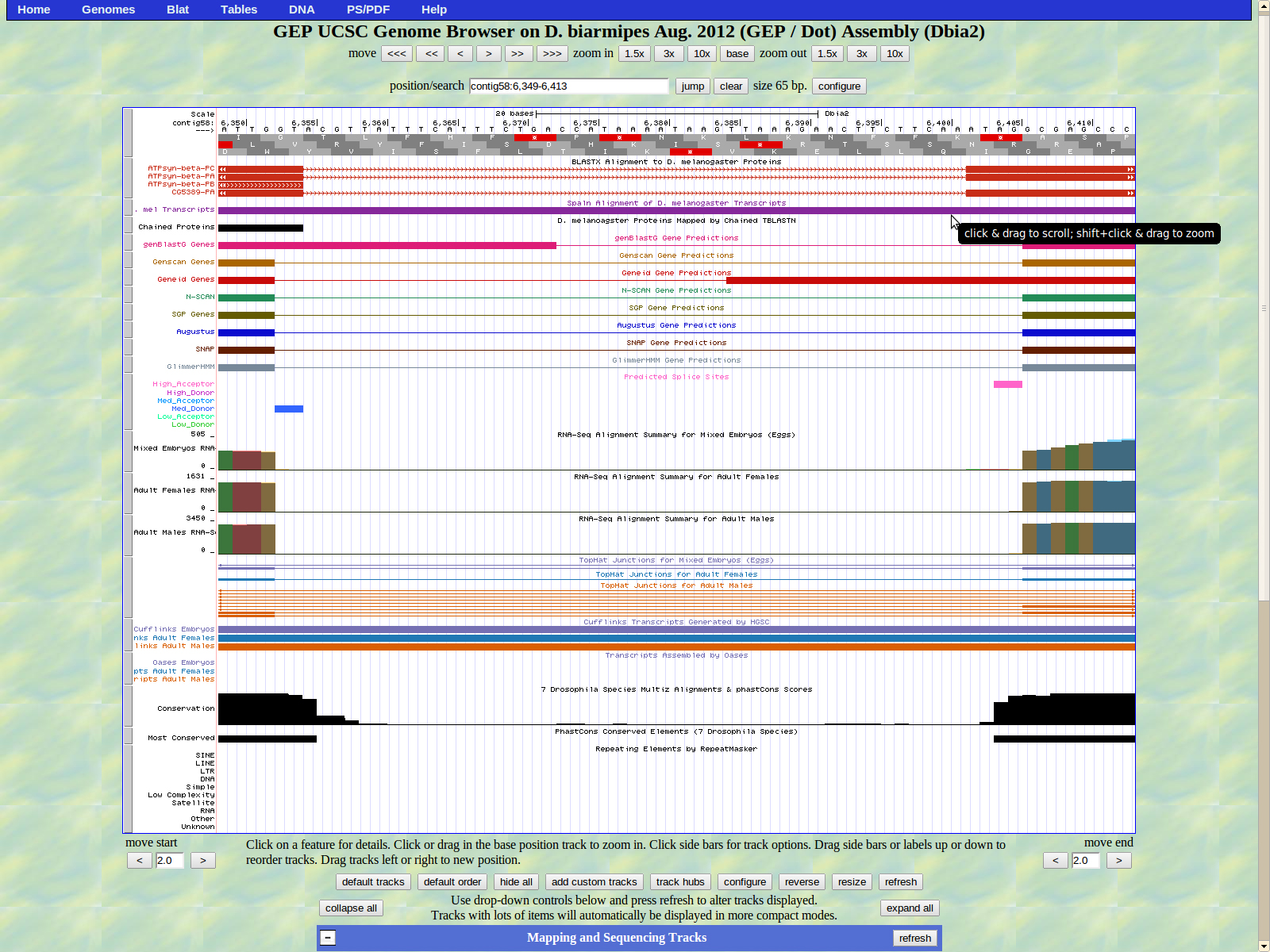
The last exon of ATPsyn-Beta ends at 7544. The stop codon TAA is located at 7545-7547 in frame +3. The GENSCAN predictors listed 7547 as the last base, which was incorrect when put into the Gene Model Checker.
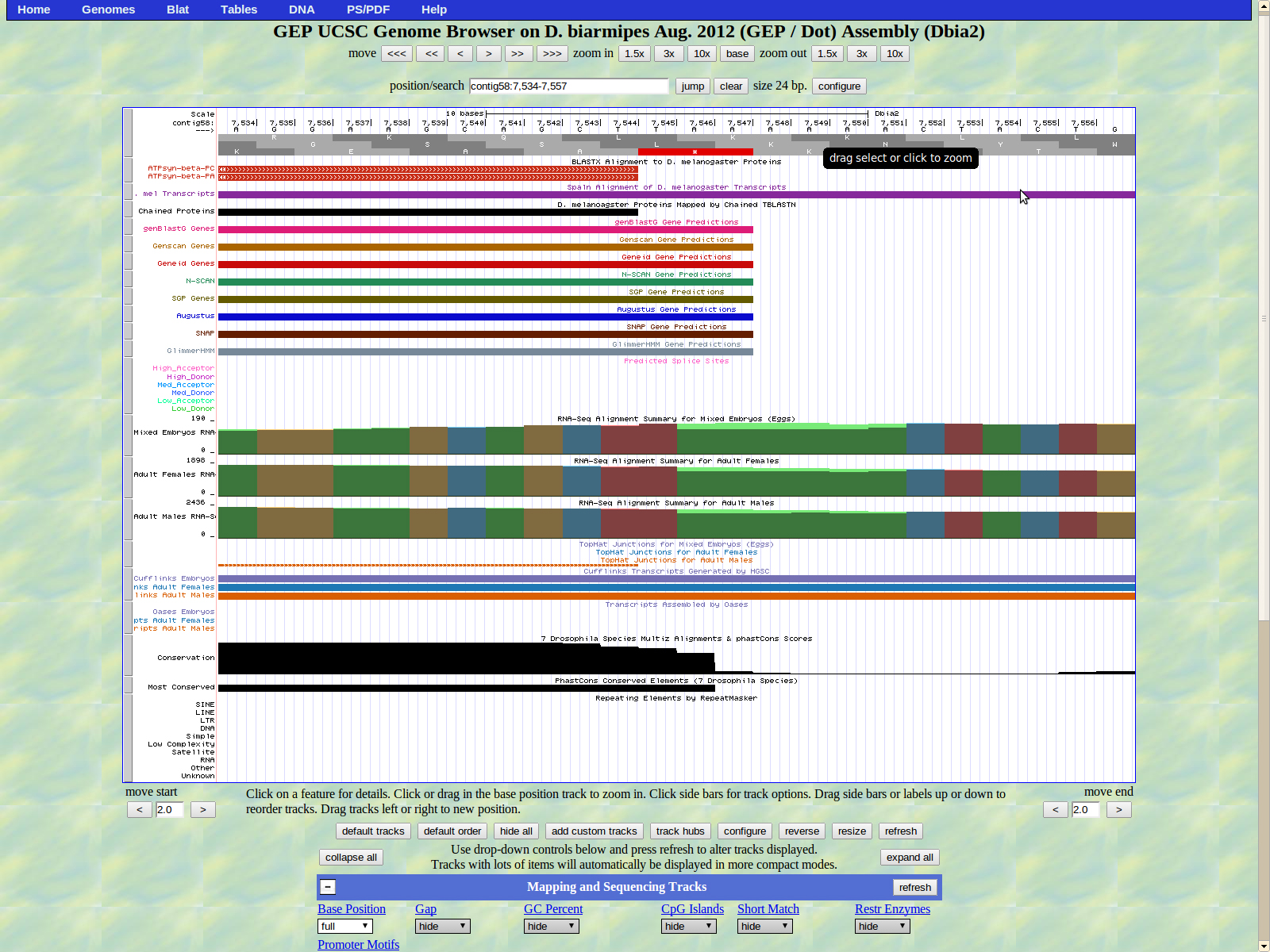
5202-5265, 6041-6352, 6406-7544 were inputed as Coding Exon Coordinates and 7545-7547 as the stop codon coordinates in the Gene Model Checker. The coordinates resulted in no checklist errors.
The Dot Matrix Alignment of D.melanogastor ATPsyn-Beta protein and the D.biarmipes ortholog displayed a nice alignment.
July 2, 2013
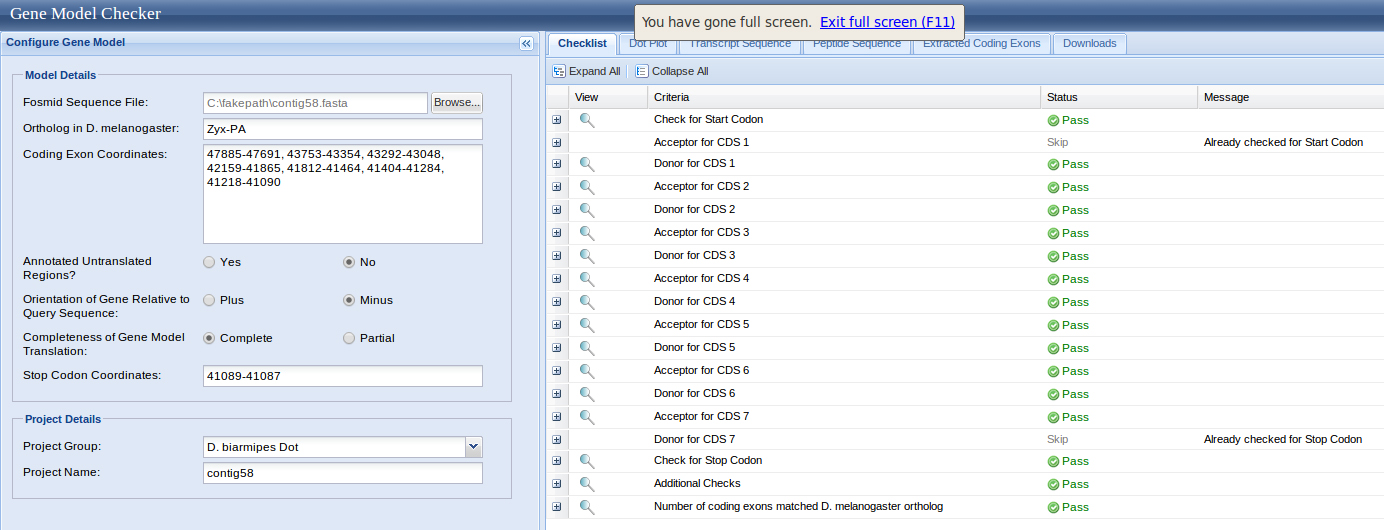
The D. biarmipes Zyxin gene is located on the negative strand of fosmid 58.
The Gene Record Finder provides:
>Zyx:11_1654_0 MQ >Zyx:8_1644_0 MESVAQQLRELSLPKGDTGSPLVCIGHGKVAKLVAKISNNQNASVKRRLD IPPKPPIKYNEMPQ >Zyx:7_1644_0 VPSSRQVLCSREPLYSQPLIGVEKTMRGHMPFRKYLSSEFGVADTQINRK TTLDNPAILEQQLEALAYHKLQMEKKGLLGVQAKPTQPLNSFTKPLSKTL SKSLIYSNLGSVRKEIETLELLTDETKISASTYSNVNET >Zyx:7_1655_0 MRGHMPFRKYLSSEFGVADTQINRKTTLDNPAILEQQLEALAYHKLQMEK KGLLGVQAKPTQPLNSFTKPLSKTLSKSLIYSNLGSVRKEIETLELLTDE TKISASTYSNVNET >Zyx:6_1655_2 MDSSHSSTQKMLSVCTNFISDNEKDELPPPPSPESAVSSSYSELRHATLE FNKPIDYLQNNQTTNPLQIYANQYAMQHDATGK >Zyx:5_1644_2 NEKDELPPPPSPESAVSSSYSELRHATLEFNKPIDYLQNNQTTNPLQIYA NQYAMQHDATGK >Zyx:4_1655_0 SSSTYDSIYEPINPRPCVADTLPRESYNLHNSYVNDNNPNISHEYNISNS IEANQTLYIHGNARTTFYDVNSIHRNDKEGLKNYISIPTEPVQELENY >Zyx:3_1655_2 RCVKCNSRVLGESSGCTAMDQIYHIFCFTCTDCQINLQGKPFYALDGKPY CEYDYLQTLEKCSVCMEPILERILRATGKPYHPQCFTCVVCGKSLDGLLF TVDATNQNYCITDFH >Zyx:2_1655_1 KFAPRCCVCKQPIMPDPGQEETIRVVALDRSFHLECYKCE >Zyx:1_1655_0 DCGLLLSSEAEGRGCYPLDDHVLCKSCNAKRVQALTNRMTSEH*
In BLASTx and aligned two or more sequences the query was my contig58.fasta and the subject was each of the above sequences. The results below were selected based on their E-value and genomic location.
The UCSC genome browser below displays the 5' end of D. biarmipes Zyx. The ATG codon (MET) spans 47885-47883 in reading frame -1 which was listed in the BLASTx results.
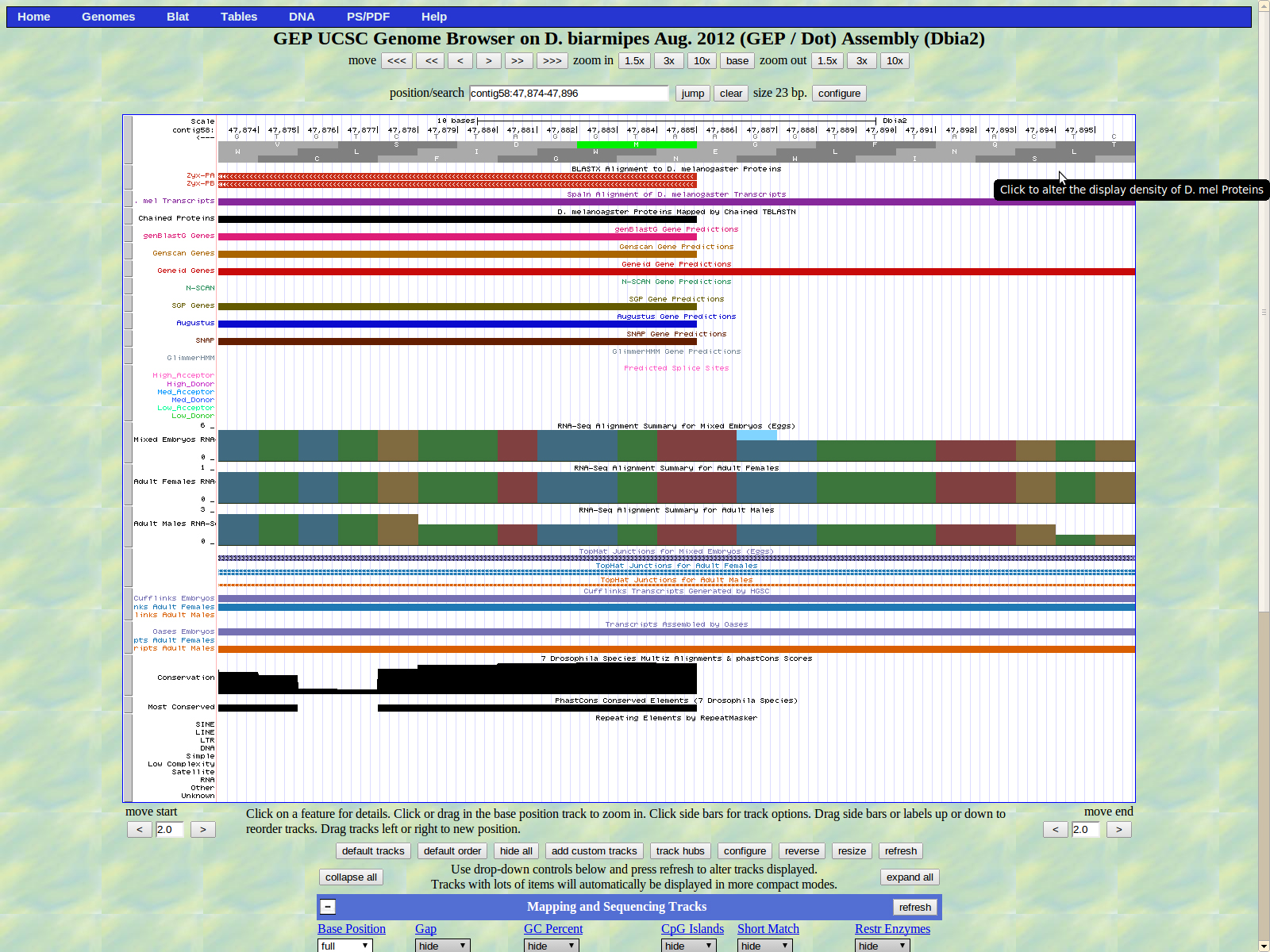
The first intron of isoforms PA and PB is displayed below. The last base of the first exon (right) is 47691. We have a medium donor GT splice site is located at 47690-47691.
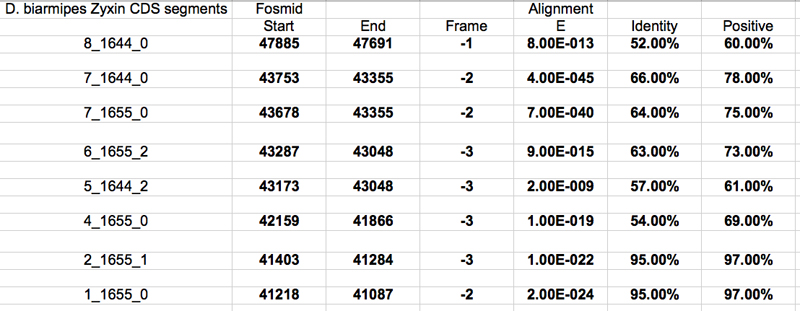
The first base of the second exon (left) is 43753; the first exon of isoforms PH and PG. The high acceptor AG splice site is located 43755-43754.
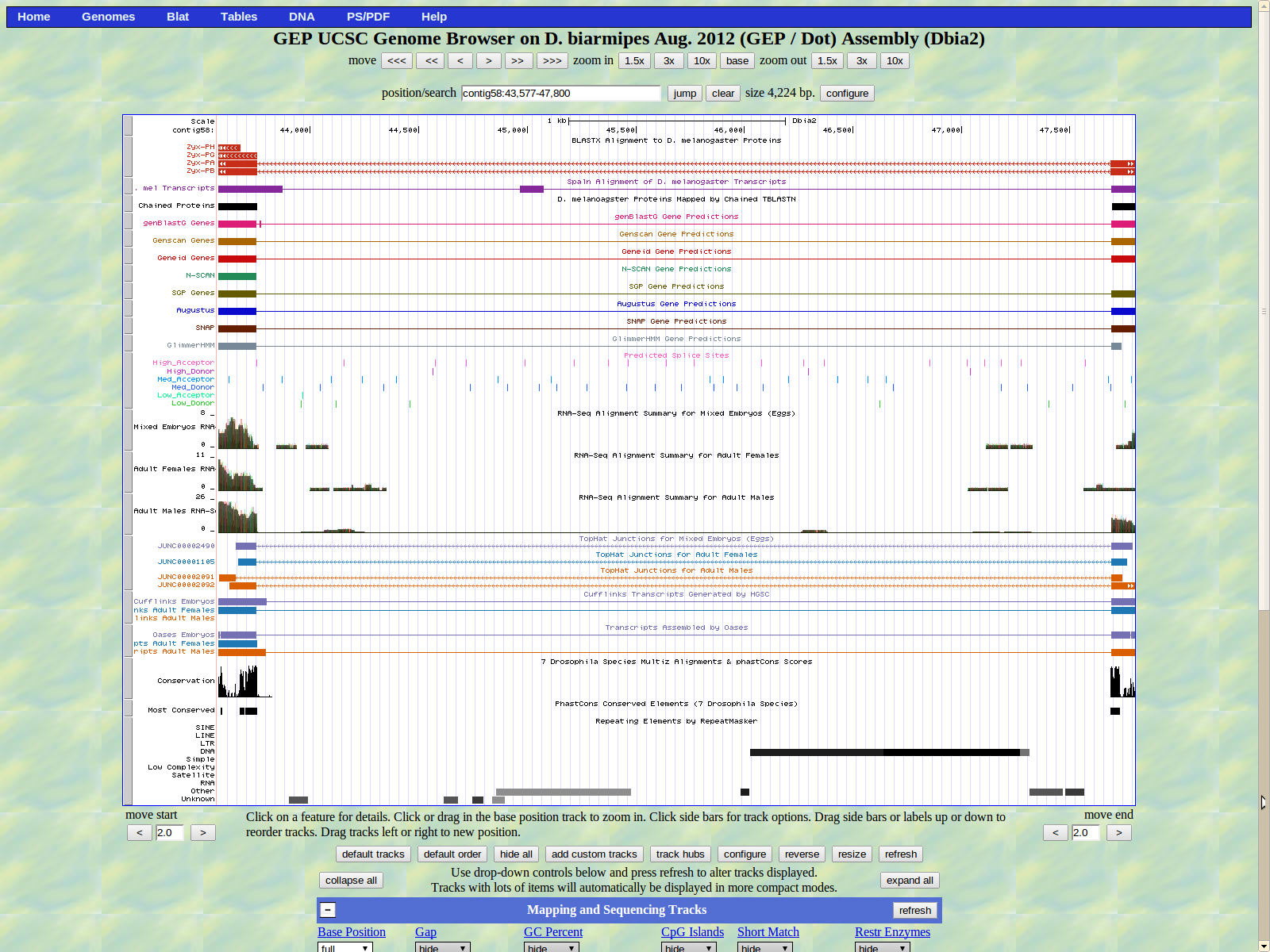
Shown below is the next intron. The last base of the previously mentioned exon (right) is 43354 in frame -2. A high donor GT splice site is shown at 43353-43352.
The next exon begins at base 43292 or 43301; both checked out as a possibility in the Gene model checker. The high acceptor splice site AG is located at 43303-43302.
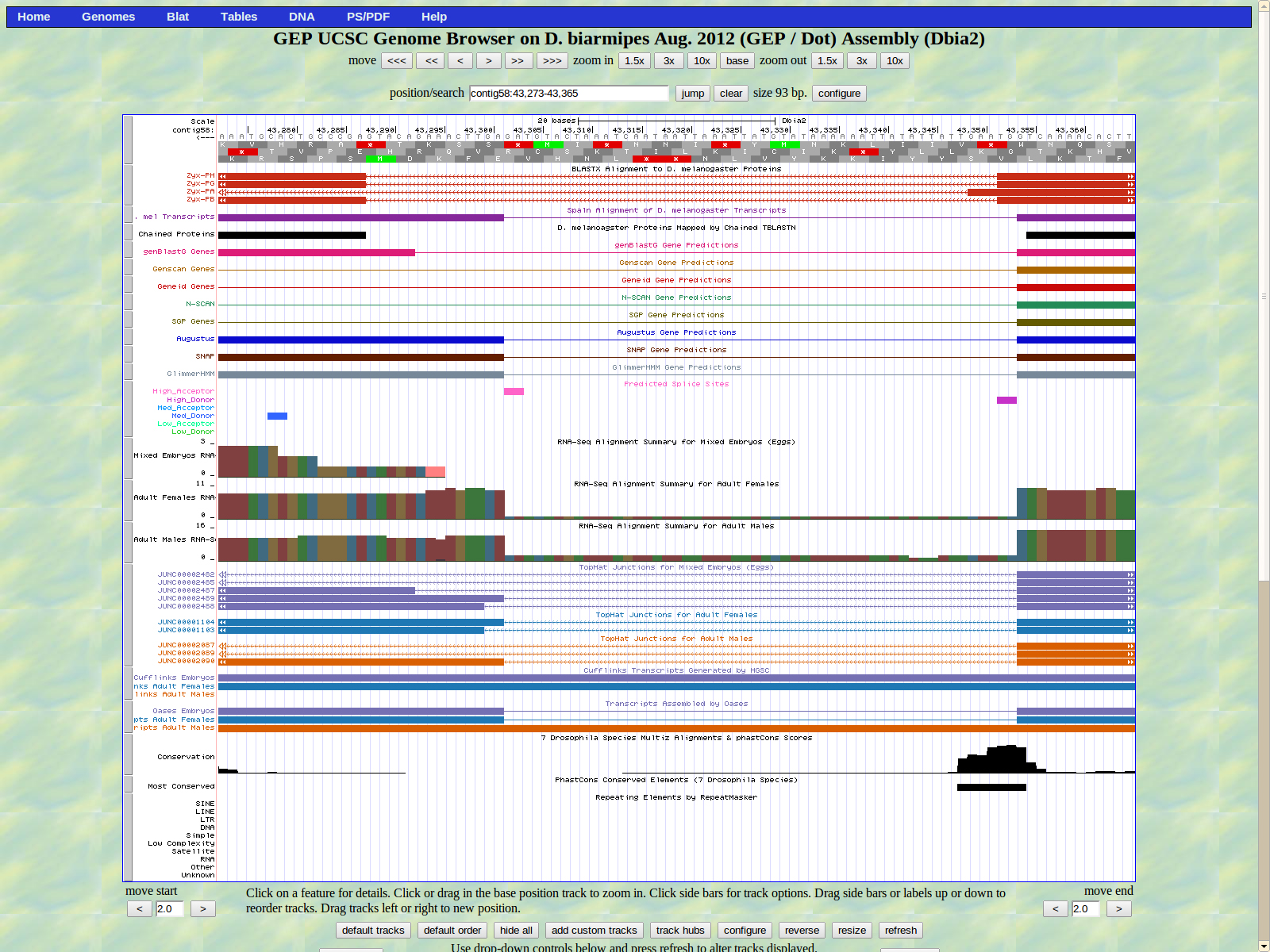
The last base of the exon (right) is 43048. The GT donor splice site is located 43047-43046 which is the medium donor prediction.
The first base of the next exon (left) is 42159. The high acceptor AG is 42161-42160.
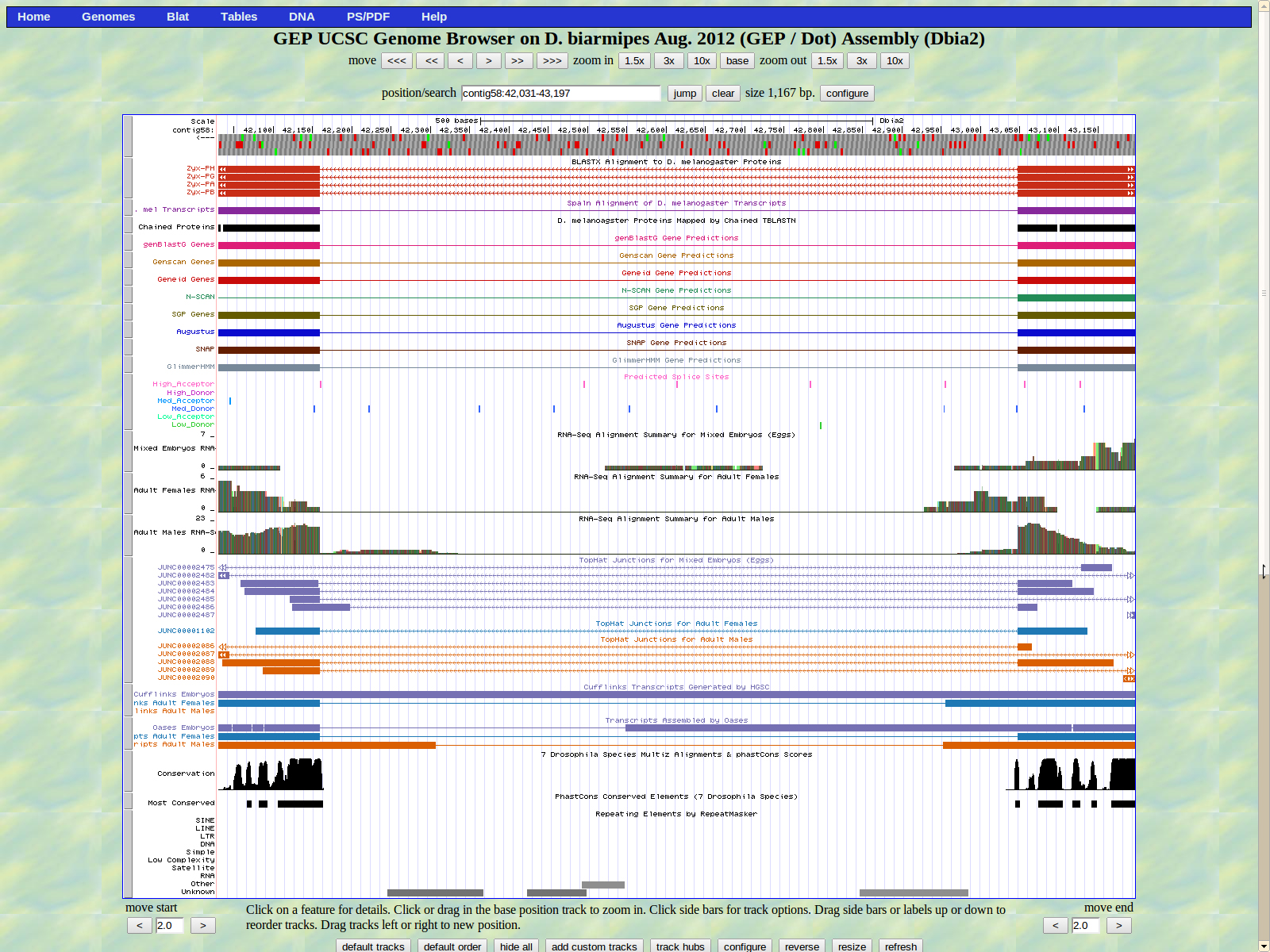
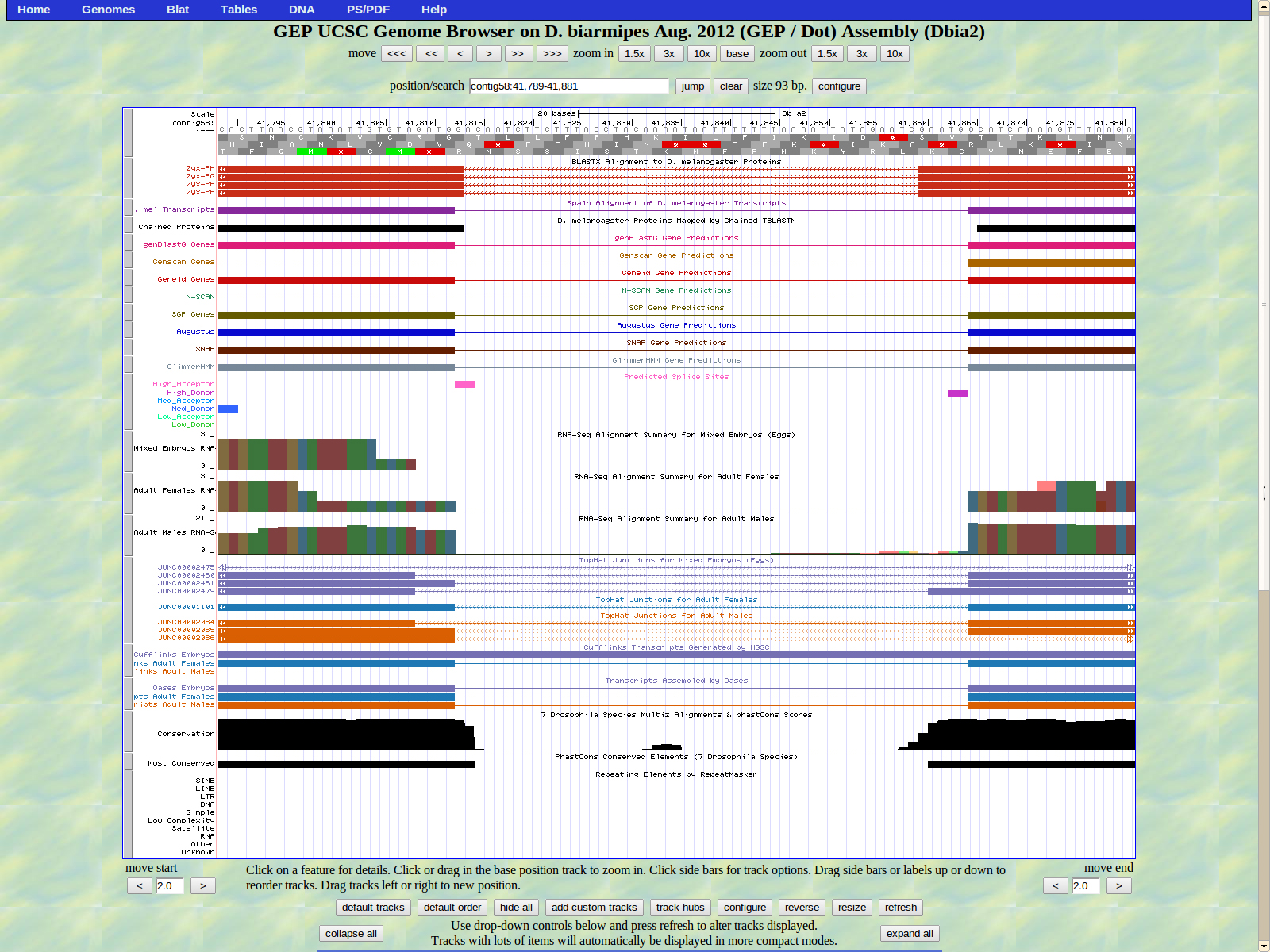
The last base of the exon (right) is 41865. The GT high donor splice site is located 41864-41863.
The first base of the next exon (left) is 41812. The high acceptor AG is 41814-41813.
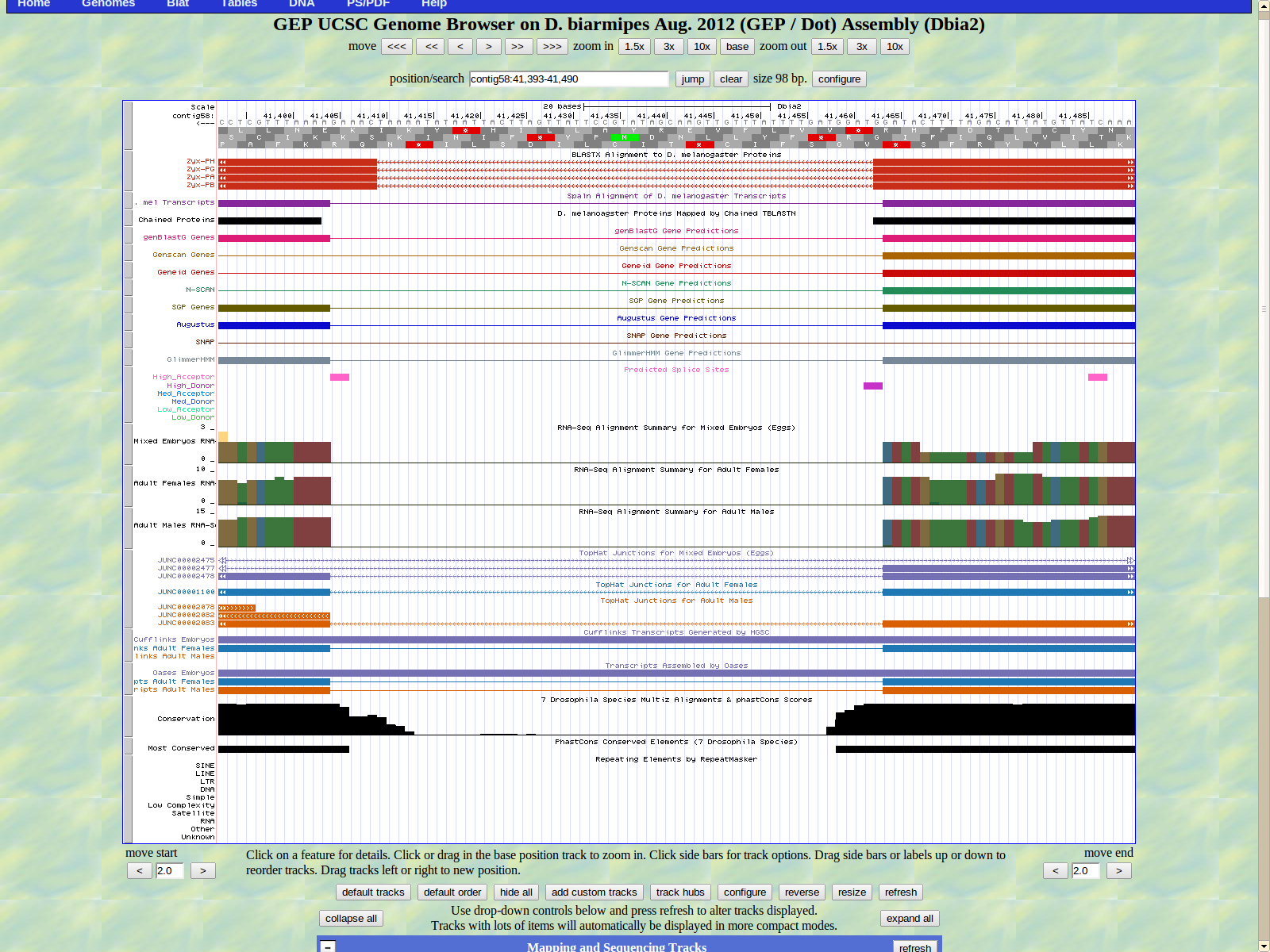
The last base of the exon (right) is 41464. The GT high donor splice site is located 41463-41462.
The first base of the next exon (left) is 41404. The high acceptor AG is 41406-414105.
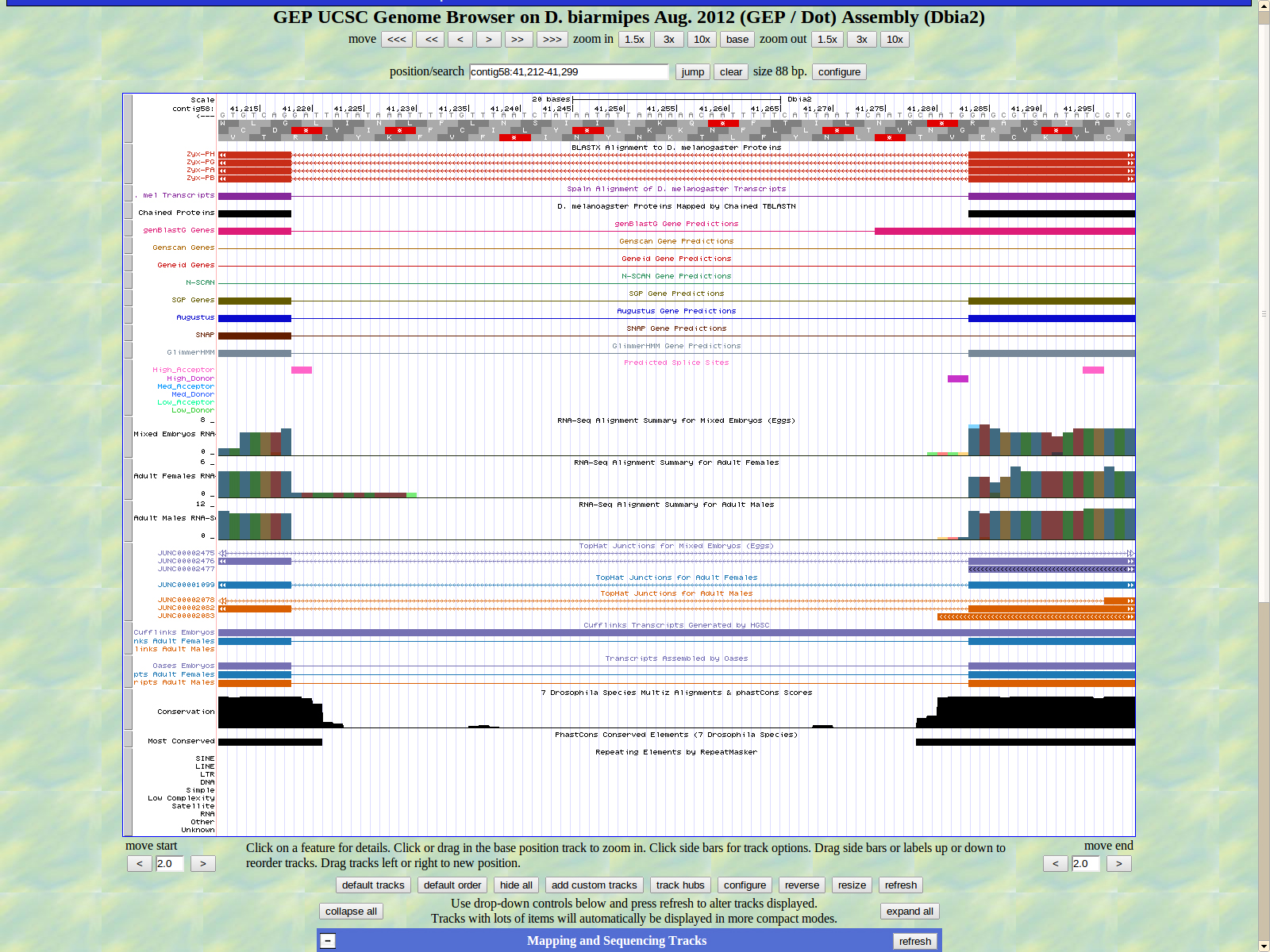
The last base of the exon (right)1284. The GT high donor splice site is located 41283-41282.
The first base of the next exon (left) is 41218. The high acceptor AG is 41220-41219.
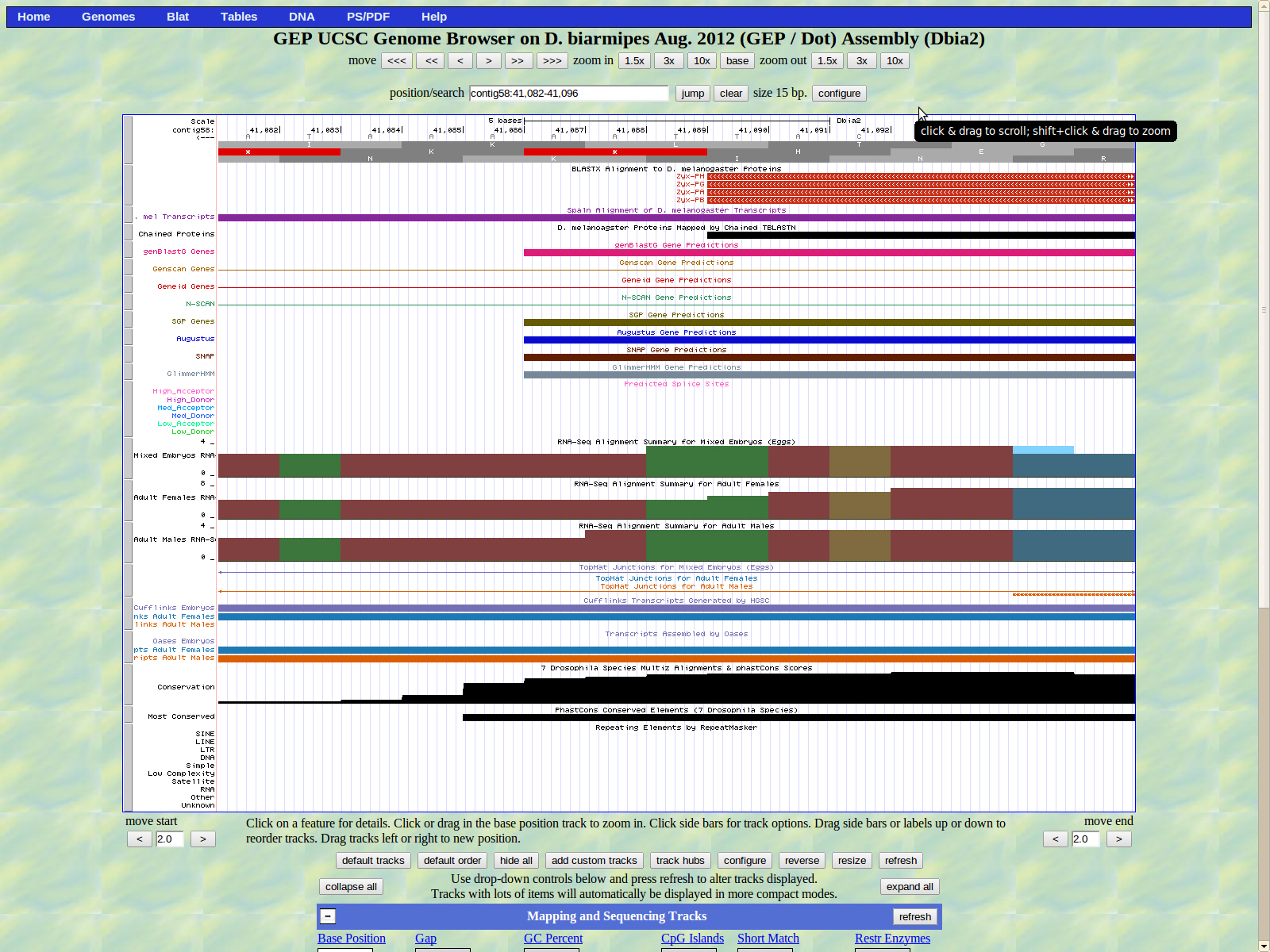
The final base of the exon is 41090. The stop codon for Zyx is 41089-41087 in from -2.
Exon coordinates 47885-47691, 43753-43354, 43292-43048 (or 43301)-43048, 42159-41865, 41812-41464, 41404-41284, 41218-41090 were inputed. The stop codon coordinates used were 41089-41087.
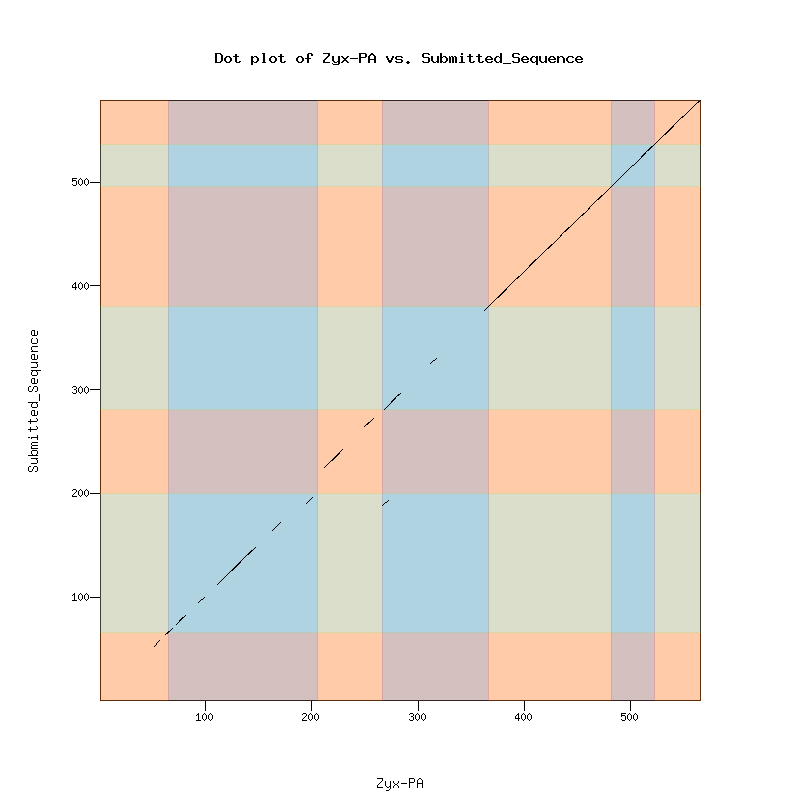
The Dot Matrix Alignment of D. melanogastor Zyxin protein and the D. biarmipes ortholog displayed a nice alignment.
A total of four isoforms as displayed is the UCSC genome browser for Zyx. PH and PG have 6 exons. PA and PB have 7 exons.
July 2, 2013
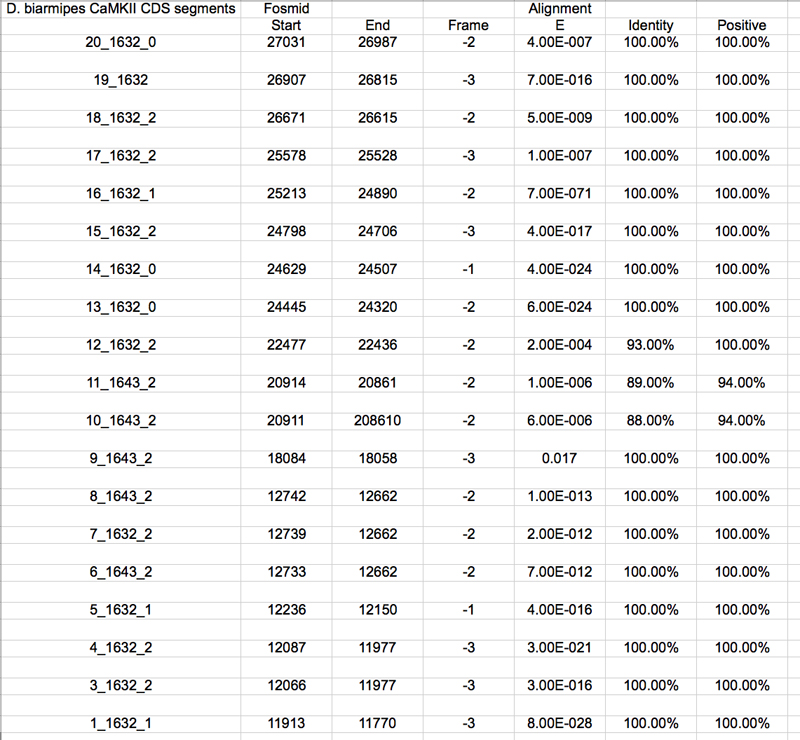
The D. biarmipes CaMKII gene is located on the negative strand of fosmid 58.
The Gene Record Finder provides:
>CaMKII:20_1632_0 MAAPAACTRFSDNYDIKEELG >CaMKII:19_1632_1 GAFSIVKRCVQKSTGFEFAAKIINTKKLTAR >CaMKII:18_1632_2 FQKLEREARICRKLHHPNI >CaMKII:17_1632_2 RLHDSIQEENYHYLVFD >CaMKII:16_1632_1 VTGGELFEDIVAREFYSEADASHCIQQILESVNHCHQNGVVHRDLKPENL LLASKAKGAAVKLADFGLAIEVQGDHQAWFGFAGTPGYLSPEVLKKEPYG KSVDIWAC >CaMKII:15_1632_2 VILYILLVGYPPFWDEDQHRLYSQIKAGAYD >CaMKII:14_1632_0 YPSPEWDTVTPEAKNLINQMLTVNPNKRITAAEALKHPWIC >CaMKII:13_1632_0 QRERVASVVHRQETVDCLKKFNARRKLKGAILTTMLATRNFS >CaMKII:12_1632_2 RSMITKKGEGSQVKESTDSSSTTLEDDDIK >CaMKII:11_1643_2 DKKGTVDRSTTVVSKEPE >CaMKII:10_1643_2 KKGTVDRSTTVVSKEPE >CaMKII:9_1643_2 IRILCPAKTYQQNIGNSQCSS >CaMKII:8_1643_2 NLFTNKAARRQEIIKITEQLIEAINSGDFDGY >CaMKII:7_1632_2 ARRQEIIKITEQLIEAINSGDFDGY >CaMKII:6_1643_2 RRQEIIKITEQLIEAINSGDFDGY >CaMKII:5_1632_1 KICDPHLTAFEPEALGNLVEGIDFHKFYFEN >CaMKII:4_1632_2 LGKNCKAINTTILNPHVHLLGEEAACIAYVRLTQYID >CaMKII:3_1632_2 INTTILNPHVHLLGEEAACIAYVRLTQYID >CaMKII:1_1632_1 QGHAHTHQSEETRVWHKRDNKWQNVHFHRSASAKISGATTFDFIPQK*
In BLASTx and aligned two or more sequences the query was my contig58.fasta and the subject was each of the above sequences. The results below were selected based on their E-value and genomic location.
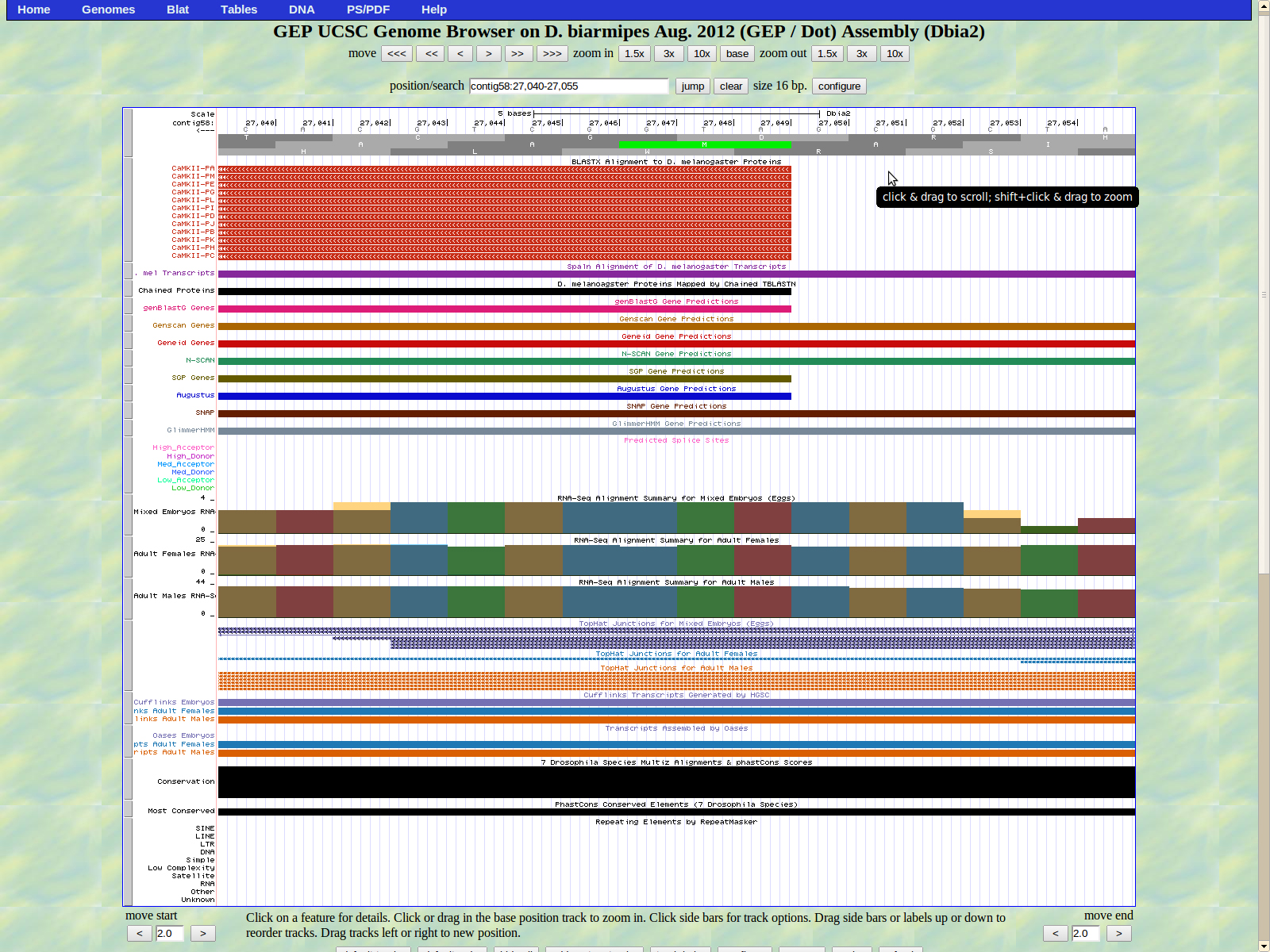
The UCSC genome browser below displays the 5' end of D. biarmipes Zyx. The ATG codon (MET) spans 27049-27047 in reading frame -2 which was listed in the BLASTx results.
The first intron of isoforms is displayed below. The last base of the first exon (right) is 26985. The high donor GT splice site is located at 26984-266983.
The first base of the second exon (left) is 26908. The high acceptor AG splice site is located 26910-26909.
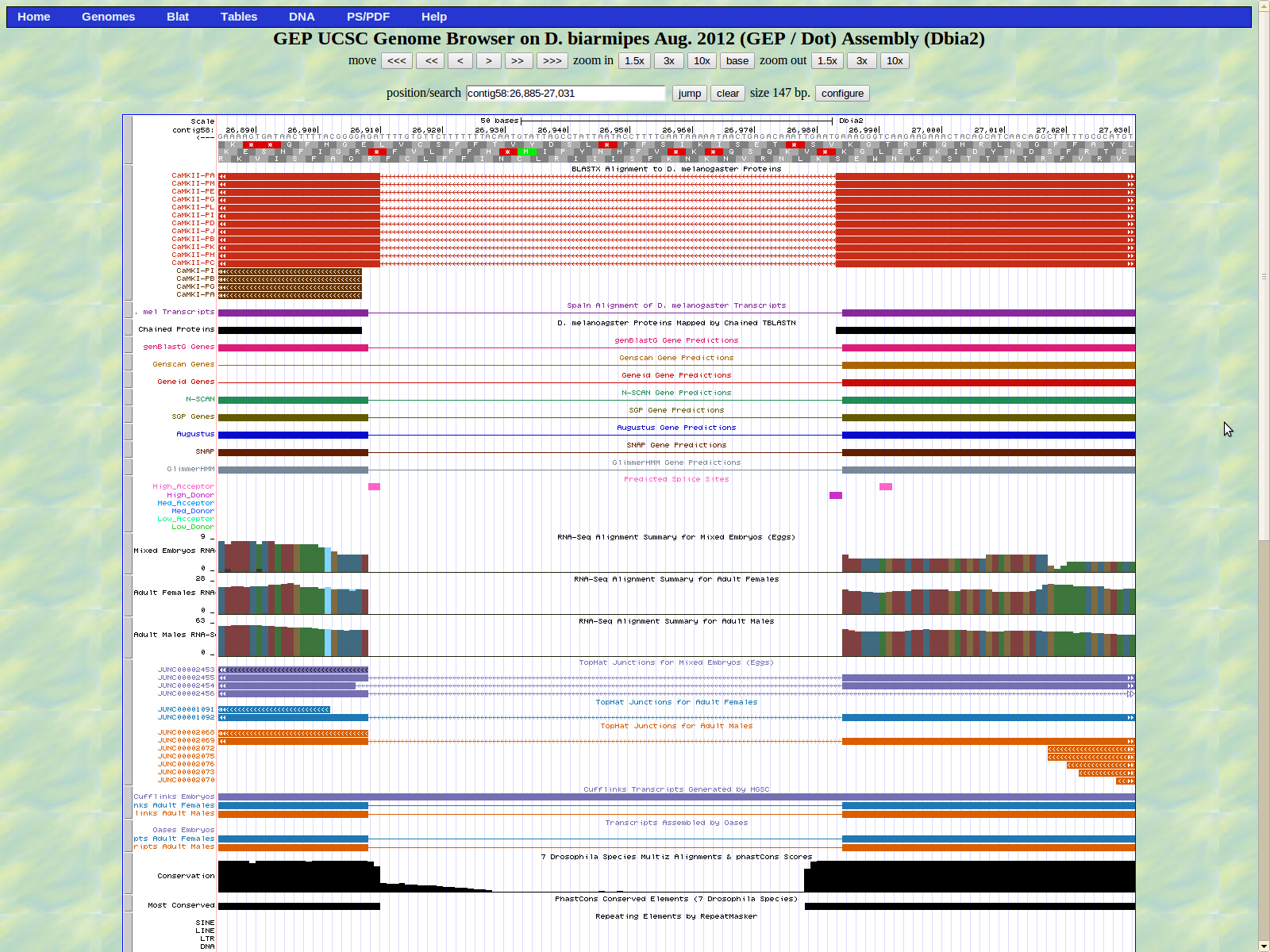
Shown below is the next intron. The last base of the previously mentioned exon (right) is 26814. A high donor GT splice site is shown at 26813-26812.
The next exon (left) begins at base 26673. The high acceptor splice site AG is located at 26728-26727 which is not proximal to the actual exon.
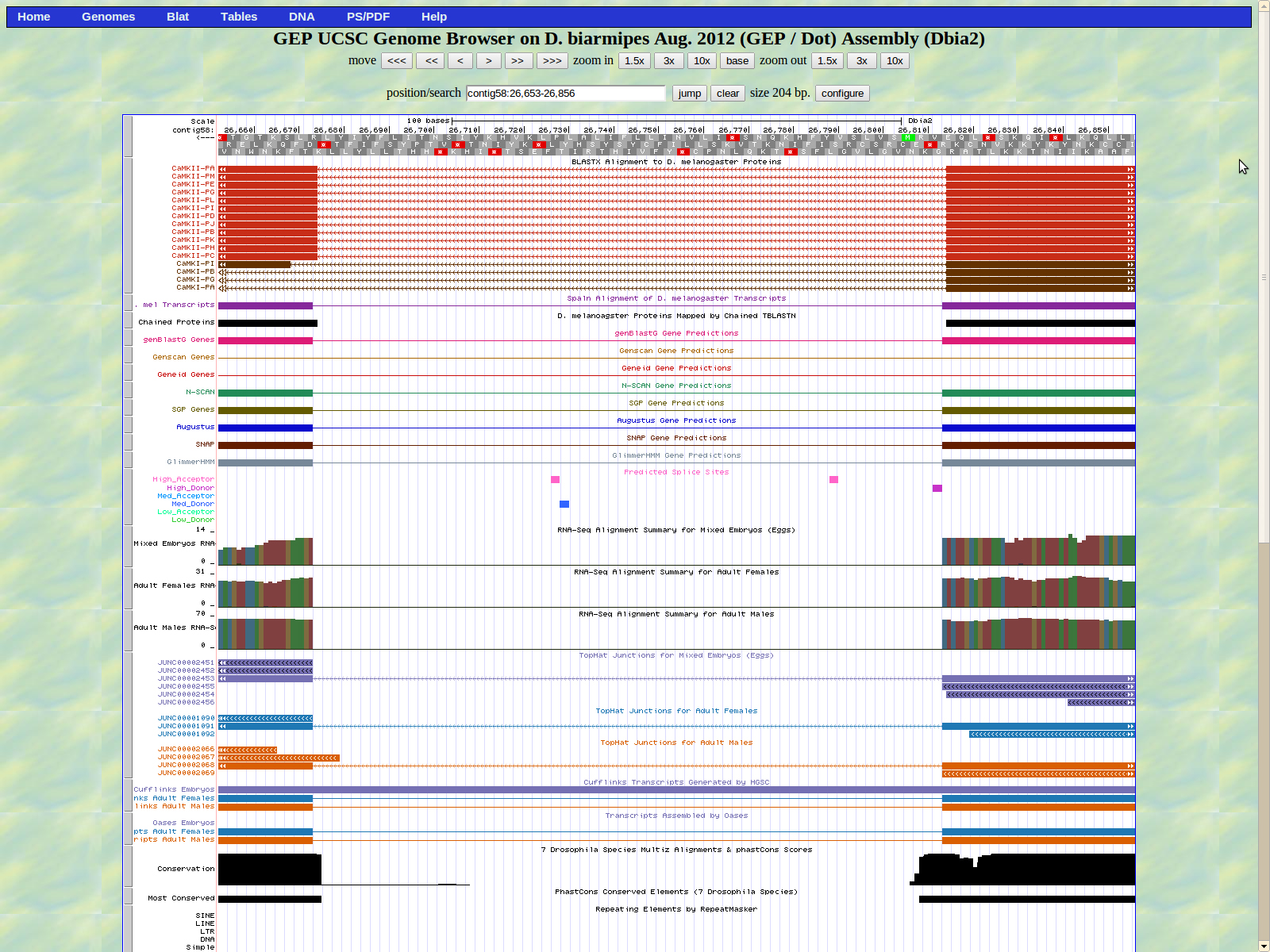
The last base of the exon (right) is 26614. The GT donor splice site is located 26613_26612 which is the medium donor prediction.
The first base of the next exon (left) is 25580. The high acceptor AG is 25582-25581.
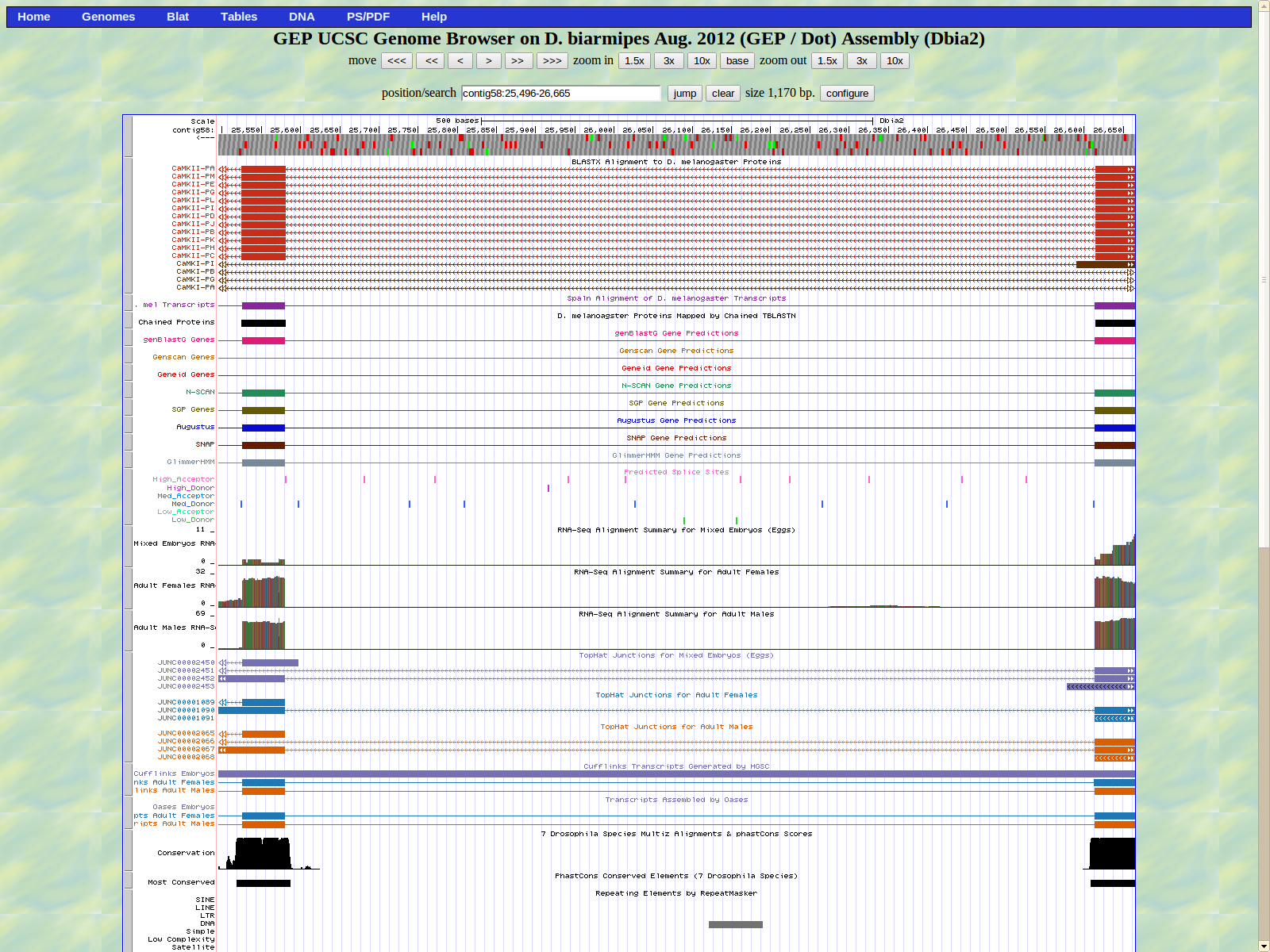
The last base of the exon (right) is 25526. The GT medium donor splice site is located 25525-25524.
The first base of the next exon (left) is 25214. The high acceptor AG is 25215-25216.

The last base of the exon (right) is. 24889 The GT high donor splice site is located 24888-24887.
The first base of the next exon (left) is 24800. The high acceptor AG is 24802-24801.

The last base of the exon (right) 24706. The GT high donor splice site is located 24705-24704.
The first base of the next exon (left) is 24629. The high acceptor AG is 24631-24630
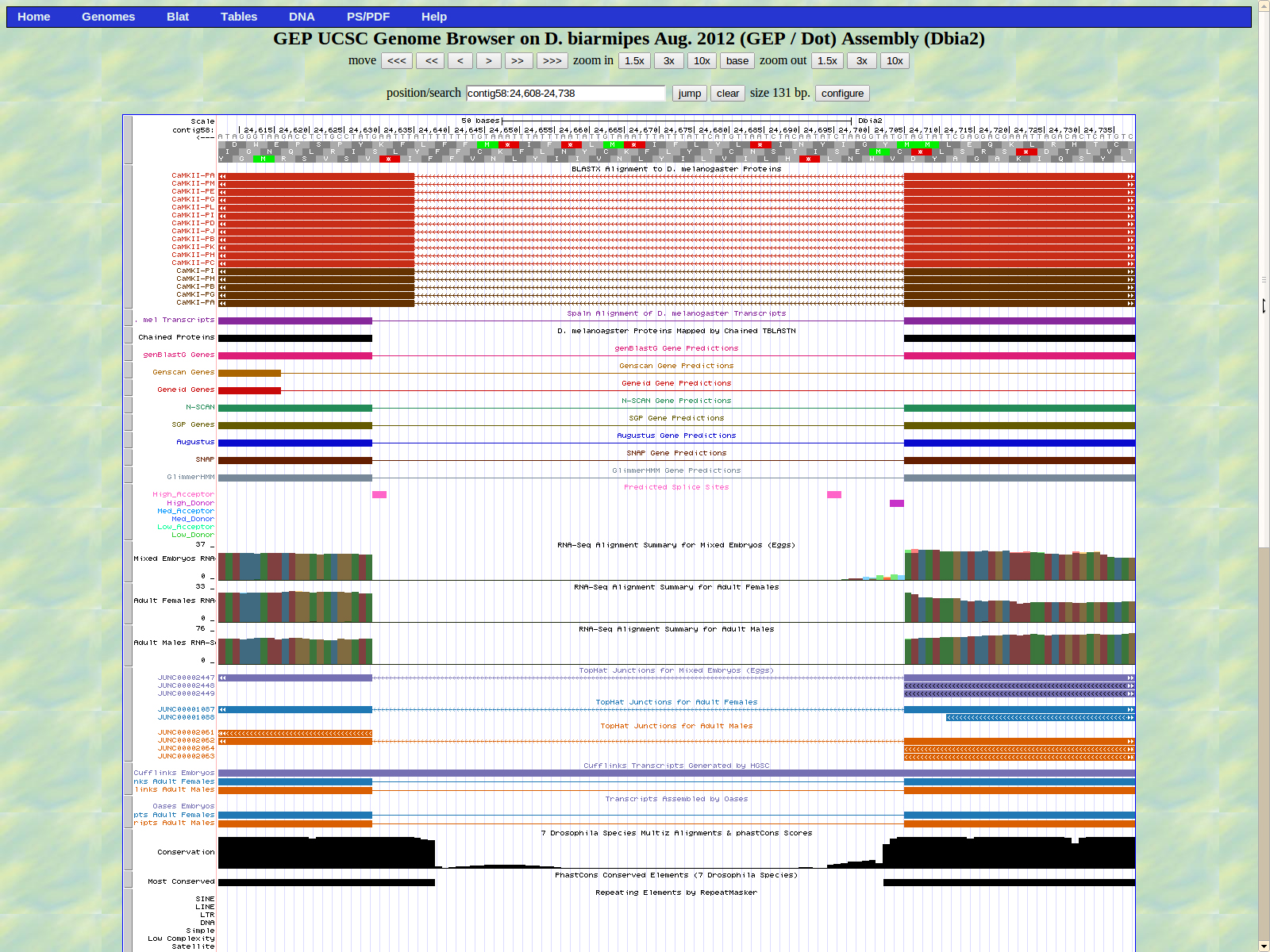
The last base of the exon (right) 24507. The GT high donor splice site is located 24506-24505.
The first base of the next exon (left) is 24445. The high acceptor AG is 24446-24447
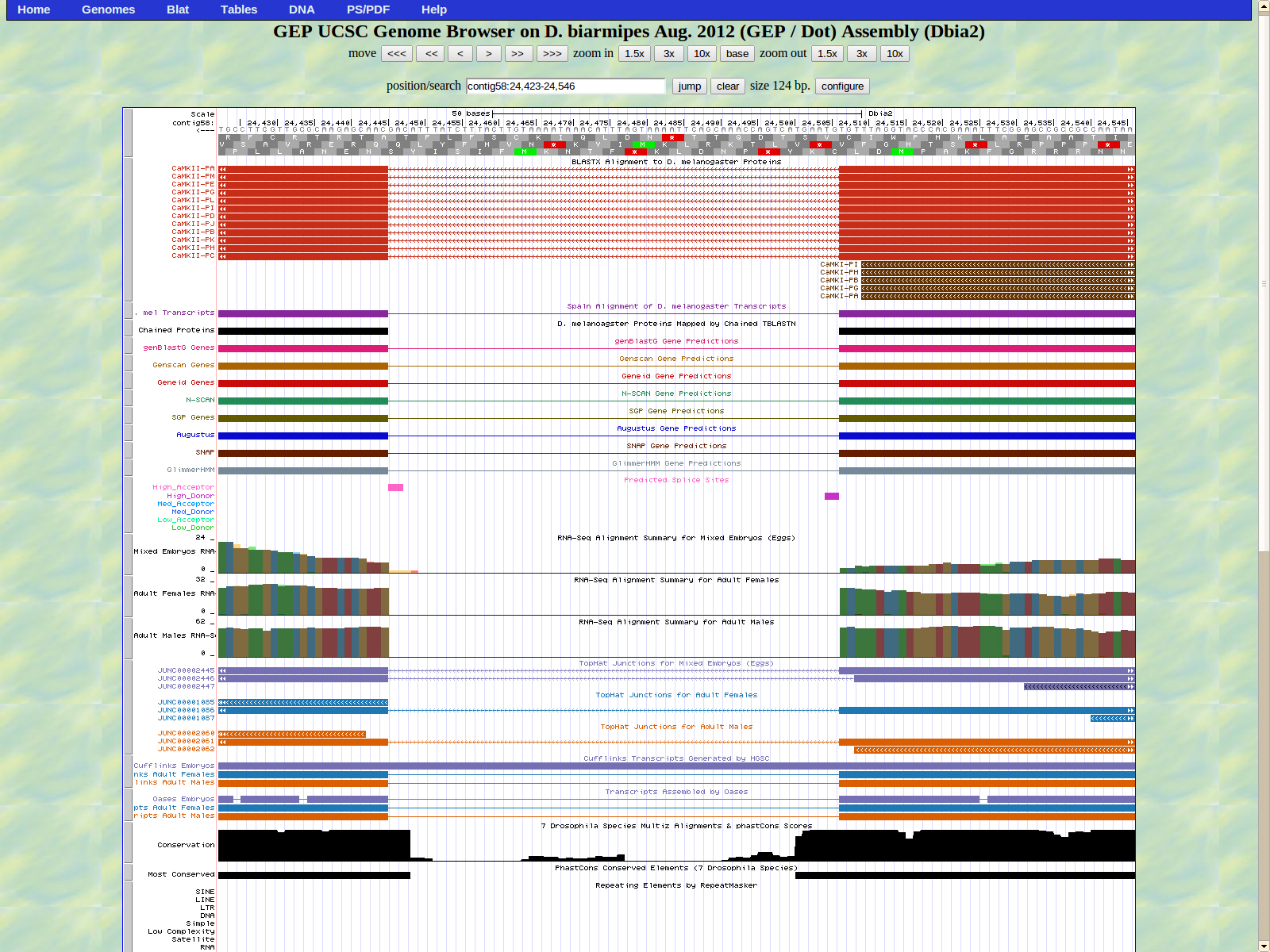
The last base of the exon (right) 24319. The GT high donor splice site is located 24318-24317.
The first base of the next exon (left) is 22479. The high acceptor AG is 22480-24481.
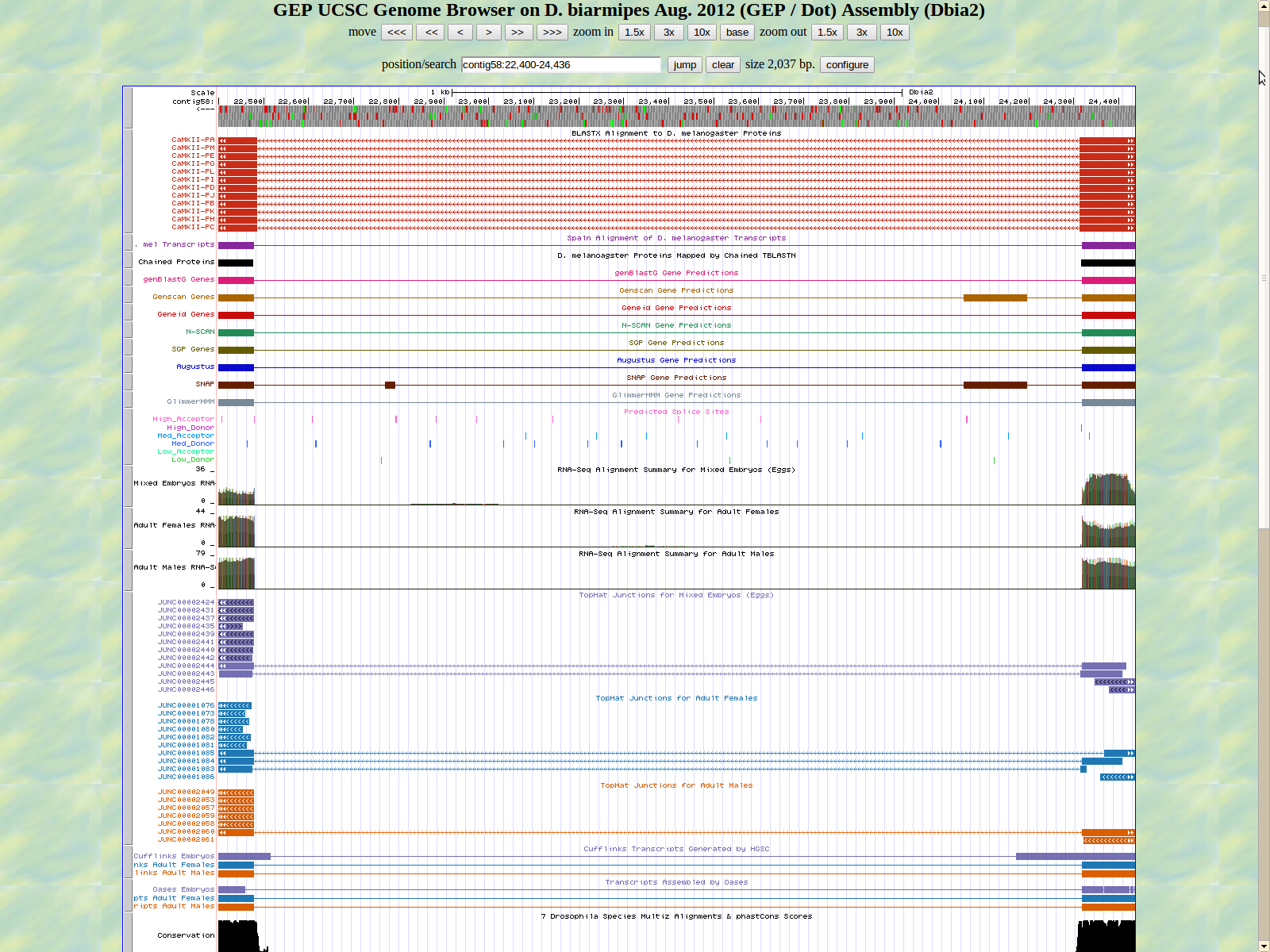
The last base of the exon (right) 22387. The GT high donor splice site is located 22386-22385.
The first base of the next exon (left) is 20916. The high acceptor AG is 20918-20917. This exon is only displayed in seven of twelve isoforms: PE, PG, PL, PI, PB, PD, and PJ.
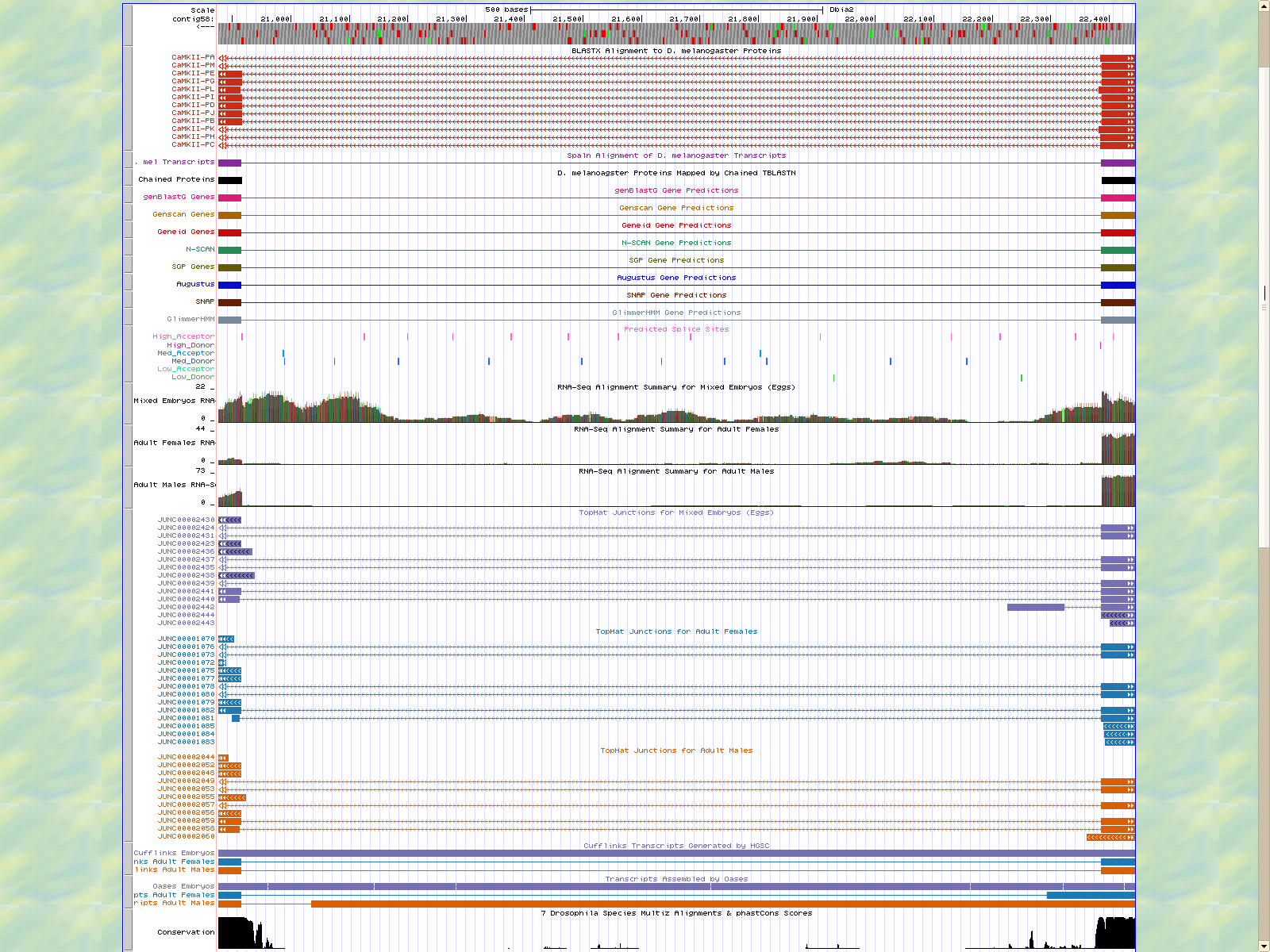
The last base of the exon (right) 20860. The GT high donor splice site is located 20859-20858.
The first base of the next exon (left) is 18086. The high acceptor AG is 18088-18087. This exon is only displayed in seven of twelve isoforms: PM, PL, PI, PD, and PJ.
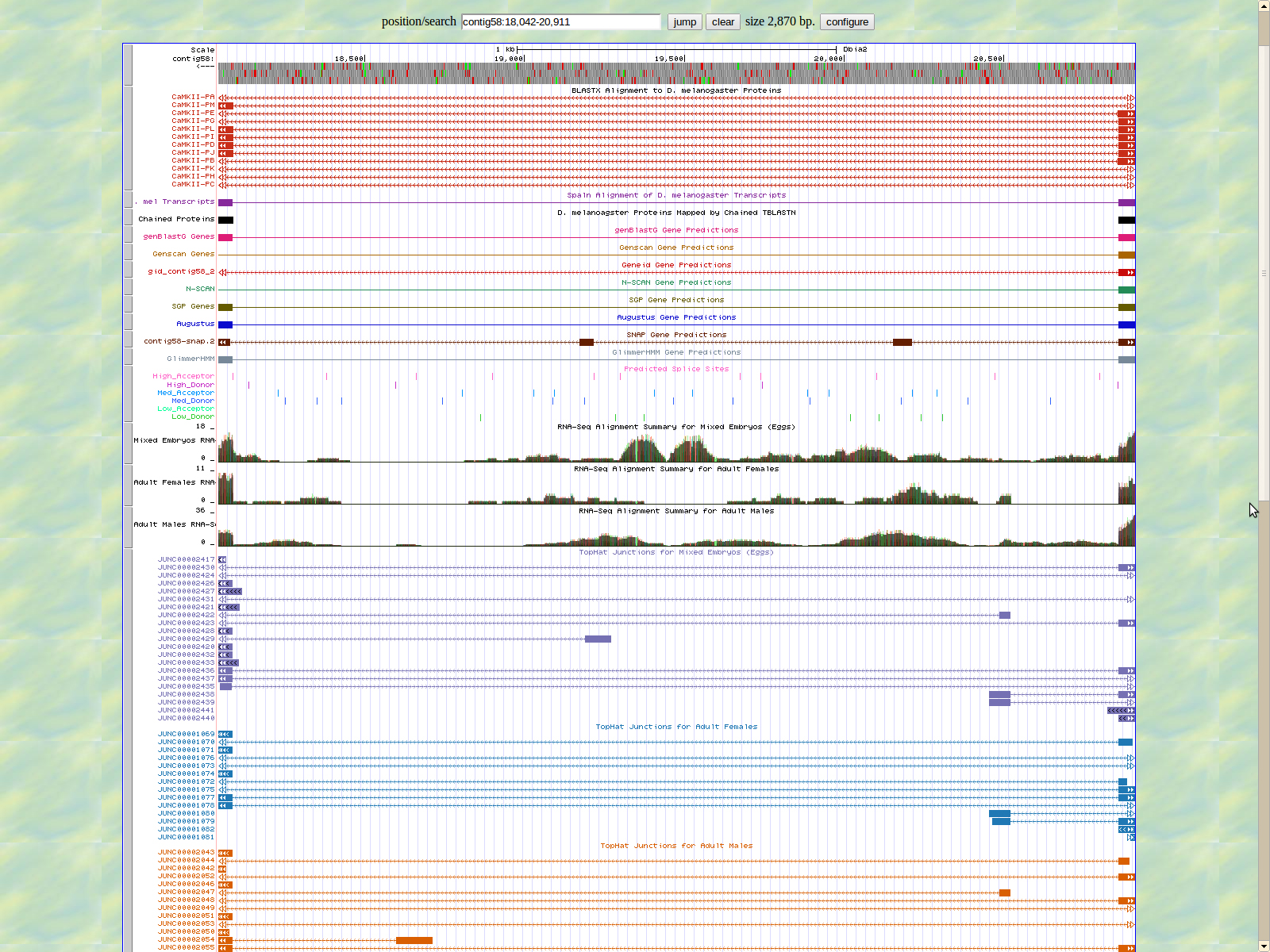
The last base of the exon (right) .18021 The GT high donor splice site is located 18020-18019.
The first base of the next exon (left) is 12738. The high acceptor AG is .12735-12734.

The last base of the exon (right) 12660. The GT high donor splice site is located 12659-12658.
The first base of the next exon (left) is 12243. The high acceptor AG is 12245-12244.
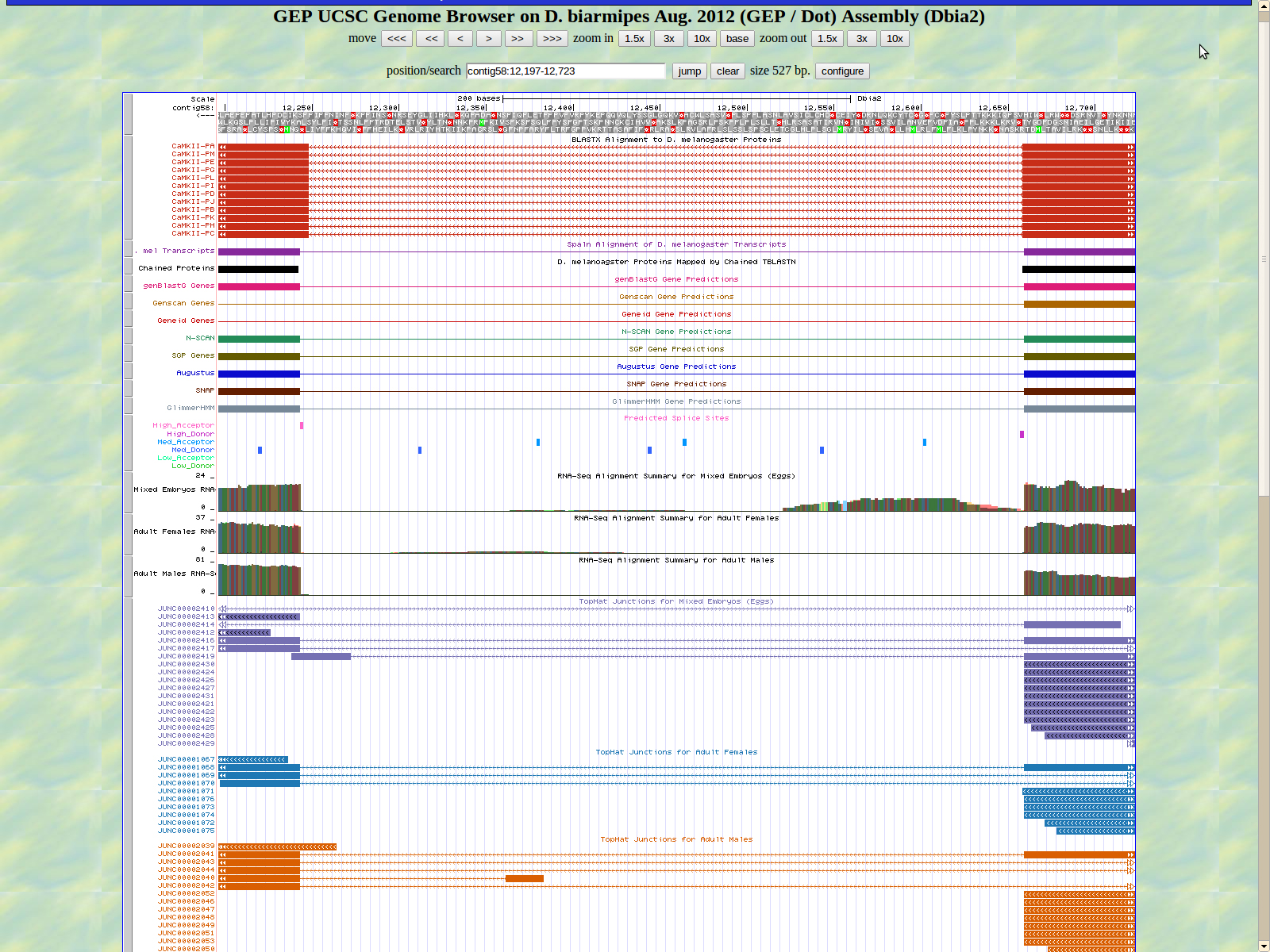
The last base of the exon (right) 12149. The GT high donor splice site is located 12148-12147.
The first base of the next exon (left) is 12089. The high acceptor AG is 12091-12090.
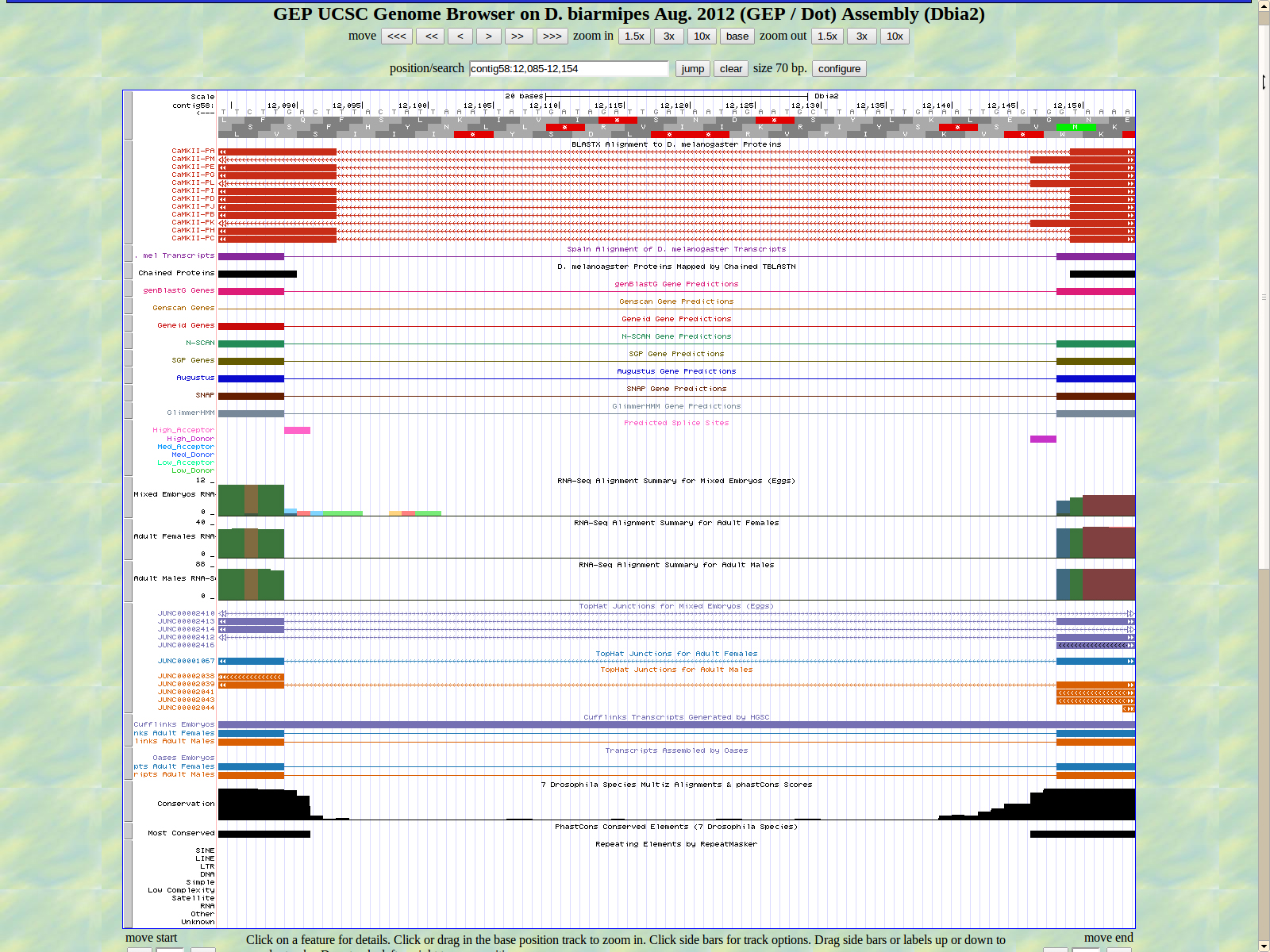
The last base of the exon (right) 11975. The GT high donor splice site is located 11974-11973.
The first base of the next exon (left) is 11914. The high acceptor AG is 11916-11915.
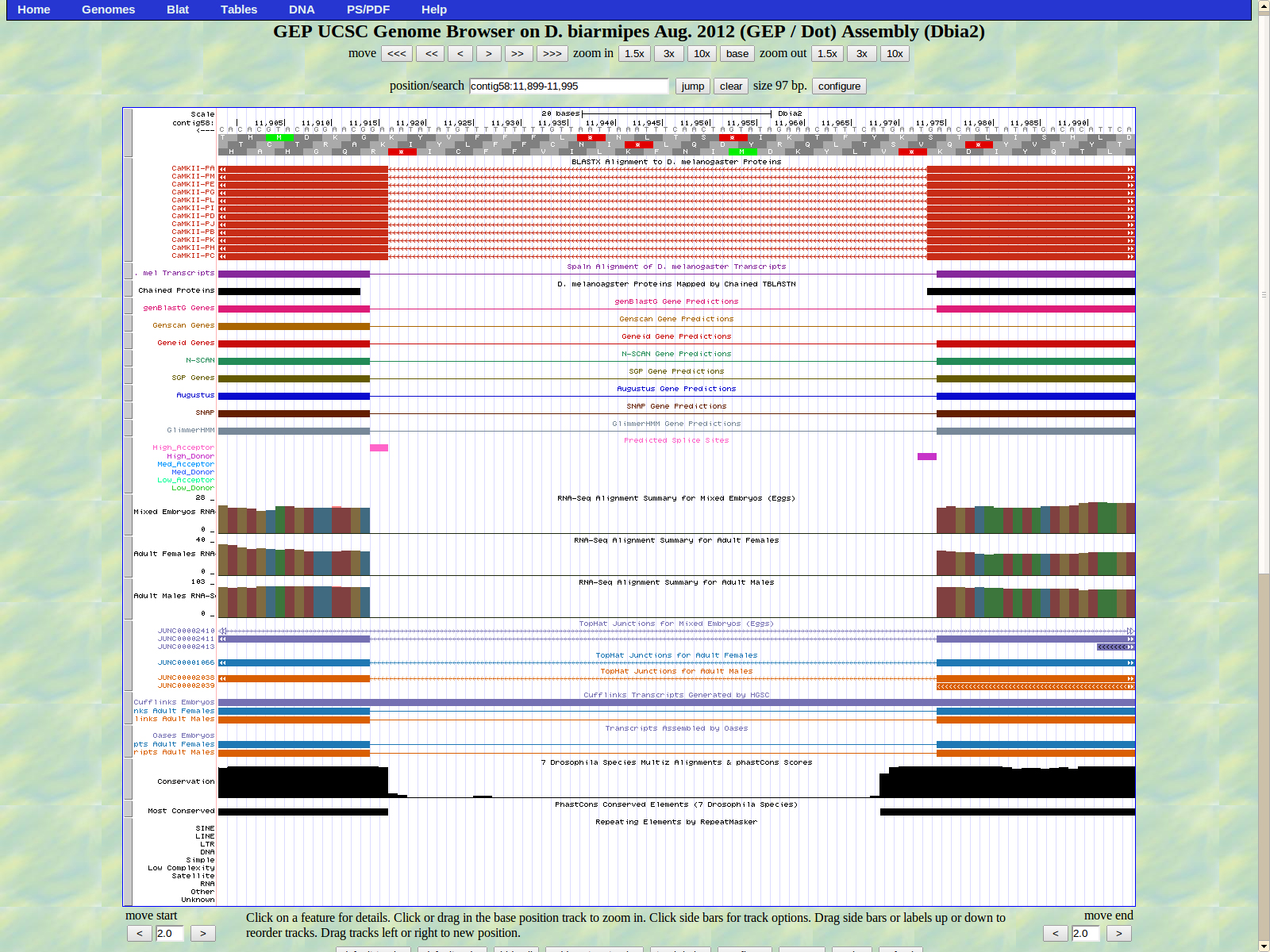
The final base of the exon is 11773. The stop codon for Zyx is 11772-11770 in frame -3.
Exon coordinates 27049-26985, 26908-26814, 26673-26614, 25580-25526, 25214 24889, 24800-24706, 24445-24319, 24479-22387, 20916-20860, 18086-18021, 12738-12660, 12243-12149, 12089-11975, 11914-11773 were inputed. The stop codon coordinates used were 11772-11770.
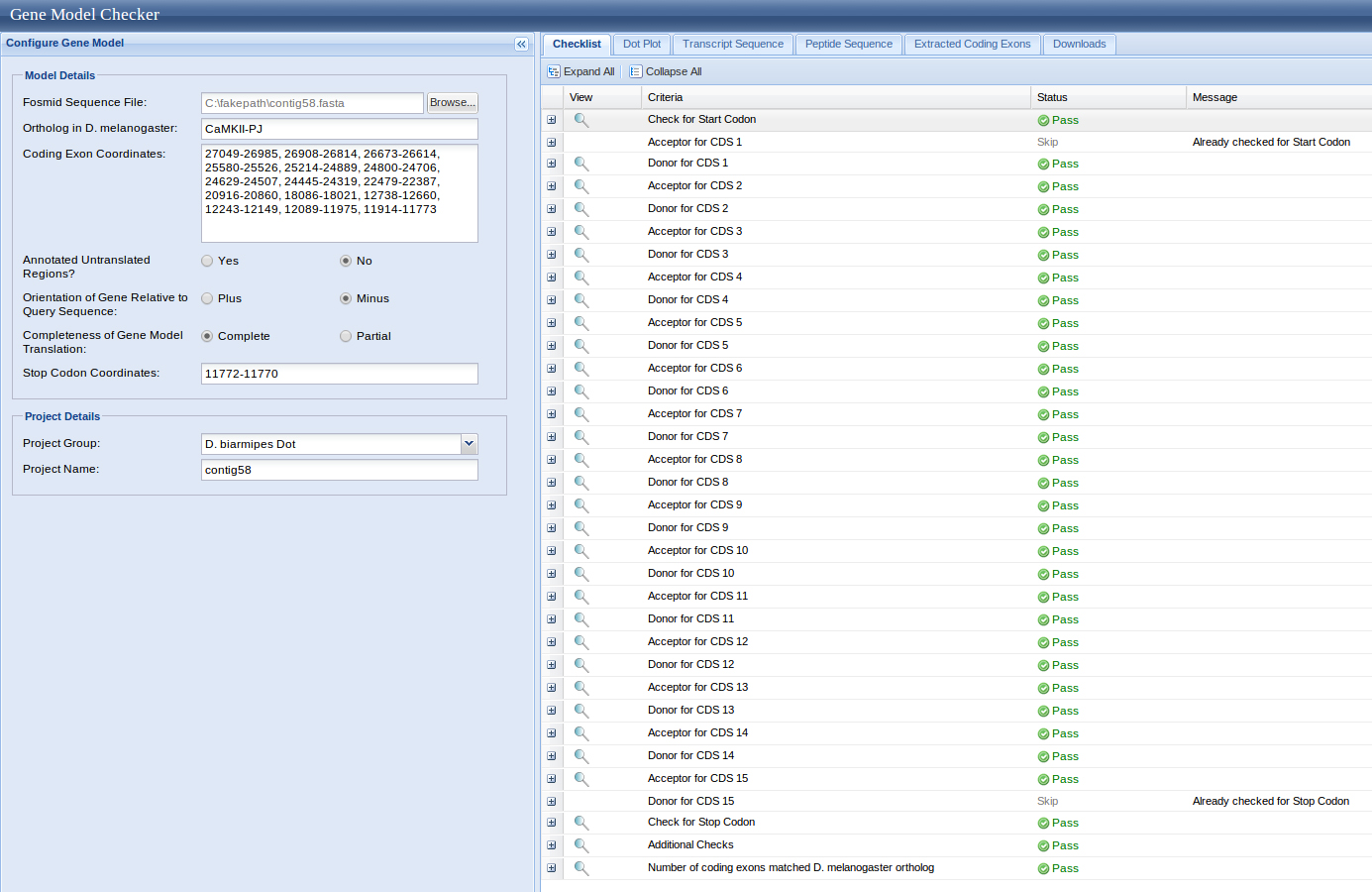
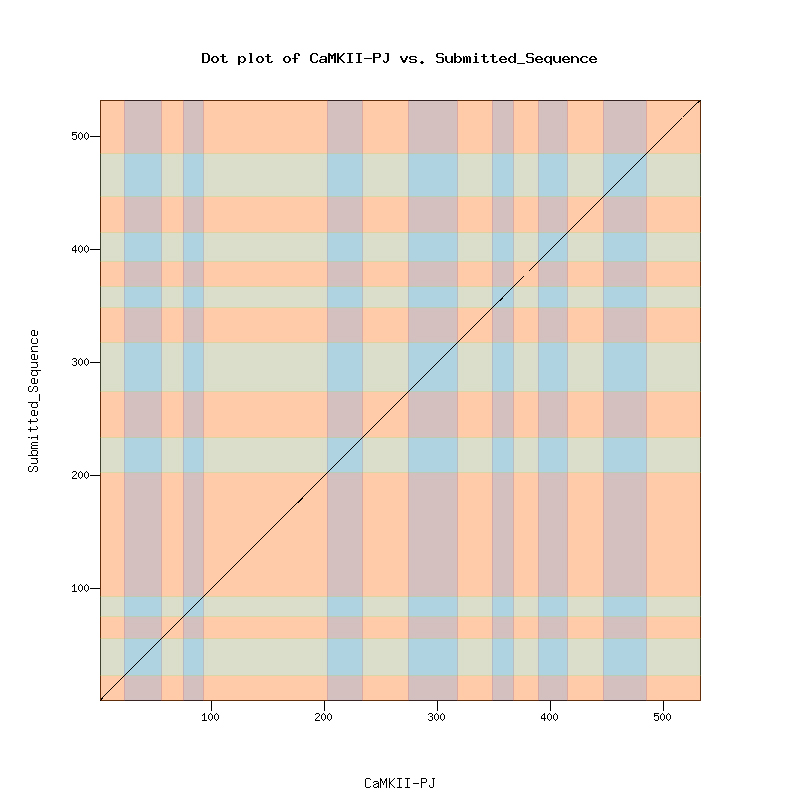
The Dot Matrix Alignment of D. melanogastor CaMKII protein and the D. biarmipes ortholog displayed a nice alignment.