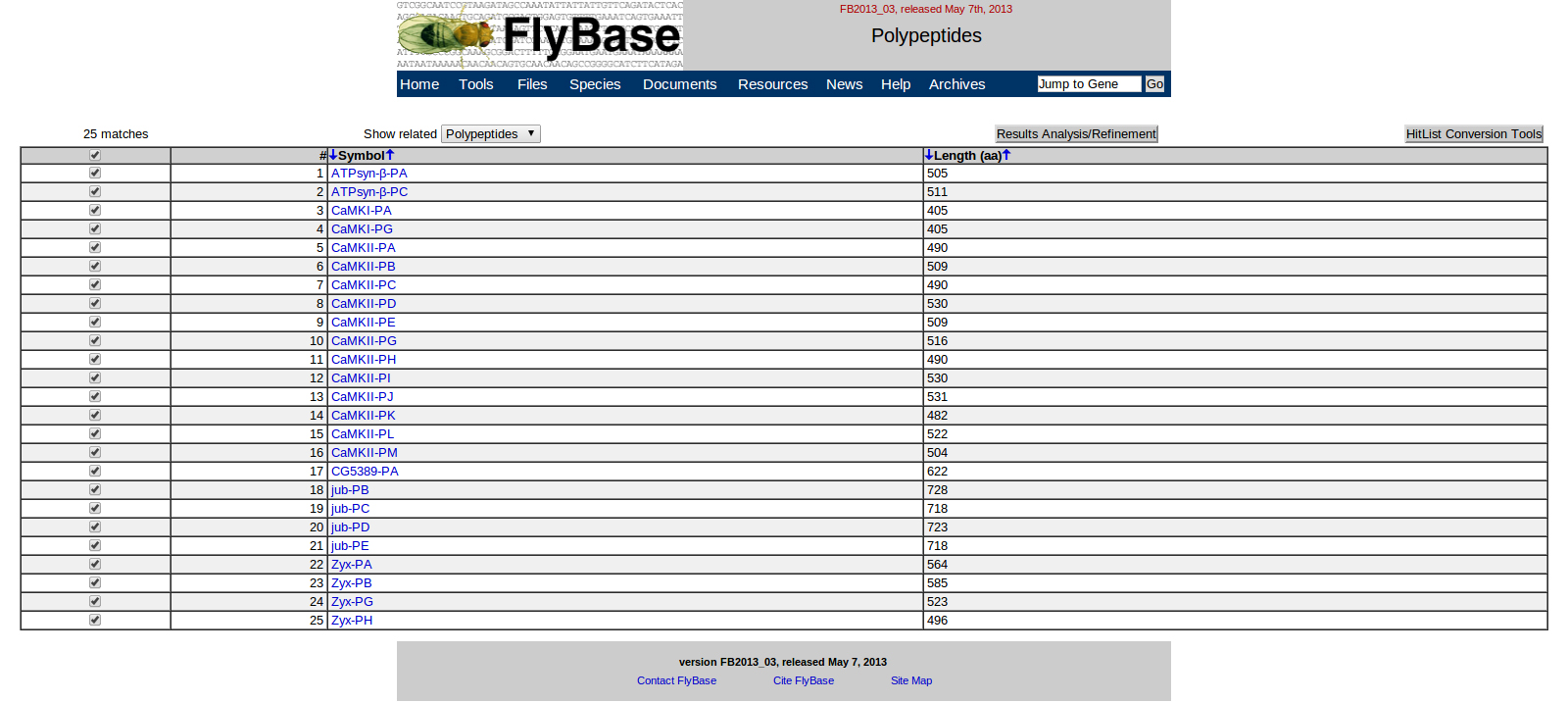
My FlyBase BLAST resulted in five gene hits: ATPsyn-beta, CaMKI, CaMKII, CG5389, jub, and Zyx.

GENSCAN results for my Contig.
>/tmp/06_18_13-07:39:46.fasta|GENSCAN_predicted_peptide_1|505_aa MFALRAASKADKNLLPFLGQLSRGHAAKAAKAAAAANGKIVAVIGAVVDVQFDDNLPPIL NALEVDNRSPRLVLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGYPIRIPVGAETLGR IINVIGEPIDERGPIETDKTAAIHAEAPEFVQMSVEQEILVTGIKVVDLLAPYAKGGKIG LFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYNEMIEGGVISLKDKTS KVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLG RIPSAVGYQPTLATDMGSMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATT VLSRAIAELGIYPAVDPLDSTSRIMDPNIIGQEHYNVARGVQKILQDYKSLQDIIAILGM DELSEEDKLTVARARKIQRFLSQPFQVAEVFTGHAGKLVPLEQTIKGFSSILAGDYDHLP EVAFYMVGPIEEVVEKADRLAKEAA >/tmp/06_18_13-07:39:46.fasta|GENSCAN_predicted_peptide_2|920_aa MDSVDQQLKELSFLKDSPSKTSSSAAIGHVNVAELVNKIRQRQSESCTMMDGPPKPPKKY NELPQVPSSKQVSCSREPLYSKPLFGIDKTMNGPMPFRKLNNSNLGIAEMSINRKTTLDN PAILEQQLEALAYHKLQMEKKGLLGVQVKASQSANSFAESPSGTLSKSLIYGNINIEKAK QTKEIGISSSTYSNVHKTEHEVEDDLLPPPSPESAVSSSYSELRQASTAFNKPTGSLQNK HKIIPSLQIYANQYELQHGAIMKSSTYDSIYEPINPRPSGDMLHRERCNLYGSSINDNNI SGSFGDVNISNSVEASHSLLYANDNCLSTCYIGKTTHKNNEKGLSNYISTAPDPIQEFEN YVPSIVLRFTSVGKLSIVIYSRRKEALRFDPSSNYAGQIKEILLYRLNTVDVHLILWTPN RRTIFQNRTDKGVPIGTVGTDFFDLASTEAFSPETWATLELAVSIRTVGSLNPVSTGTAA TNSAGESCTFVVHHRIAEPAKNKGLFRRRRKTTLKTPEKGLLGTSKRFIVAIDFLKCAIA MAAPAACTRFSDNYDIKEELGNVTGGELFEDIVAREFYSEADASHCIQQILESVNHCHQN GVVHRDLKPENLLLASKAKGAAVKLADFGLAIEVQGDHQAWFGFAGTPGYLSPEVLKKEP YGKSVDIWACEWDTVTPEAKNLINQMLTVNPNKRITAAEALKHPWICQRERVASVVHRQE TVDCLKKFNARRKLKGAILTTMLATRNFSKNTQLHGSEHIFRSCGLDISATLVYIIWCTF RFIKEIFFNAIRIAGTGRSMITKKGDGSQVKESTDSSSTTLEDDDIKEDKKGIVDRSTTV ISKEPEAARRQEIIKITEQLIEAINSGDFDGYTQGHAHTHQSEETRVWHKRDNKWQNVHF HRSATAKISGATTFDFIPQK
I ran a blastp for these two sequences. Results were three genes: ATPsyn-beta, CaMKII, and Zyxin.
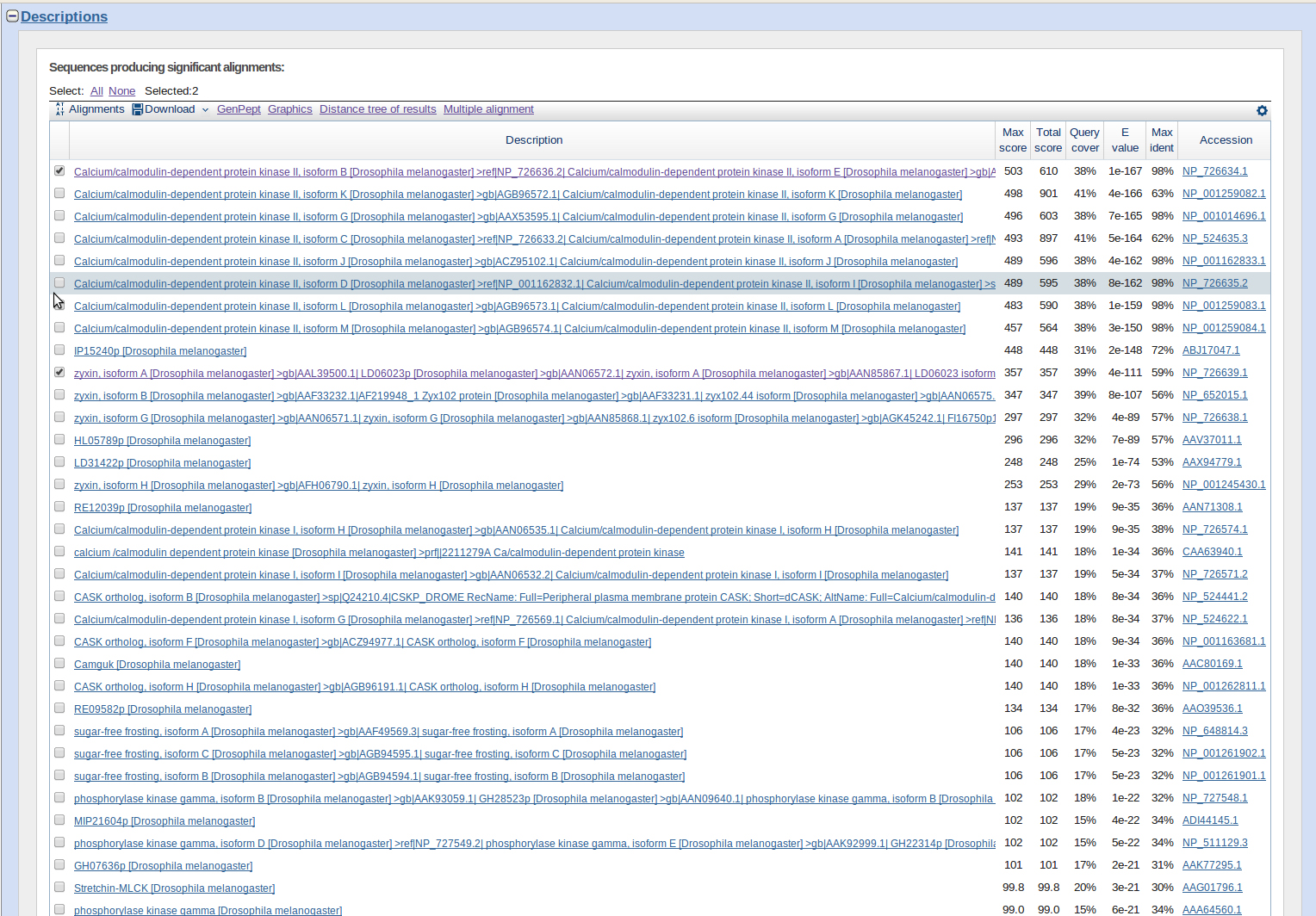
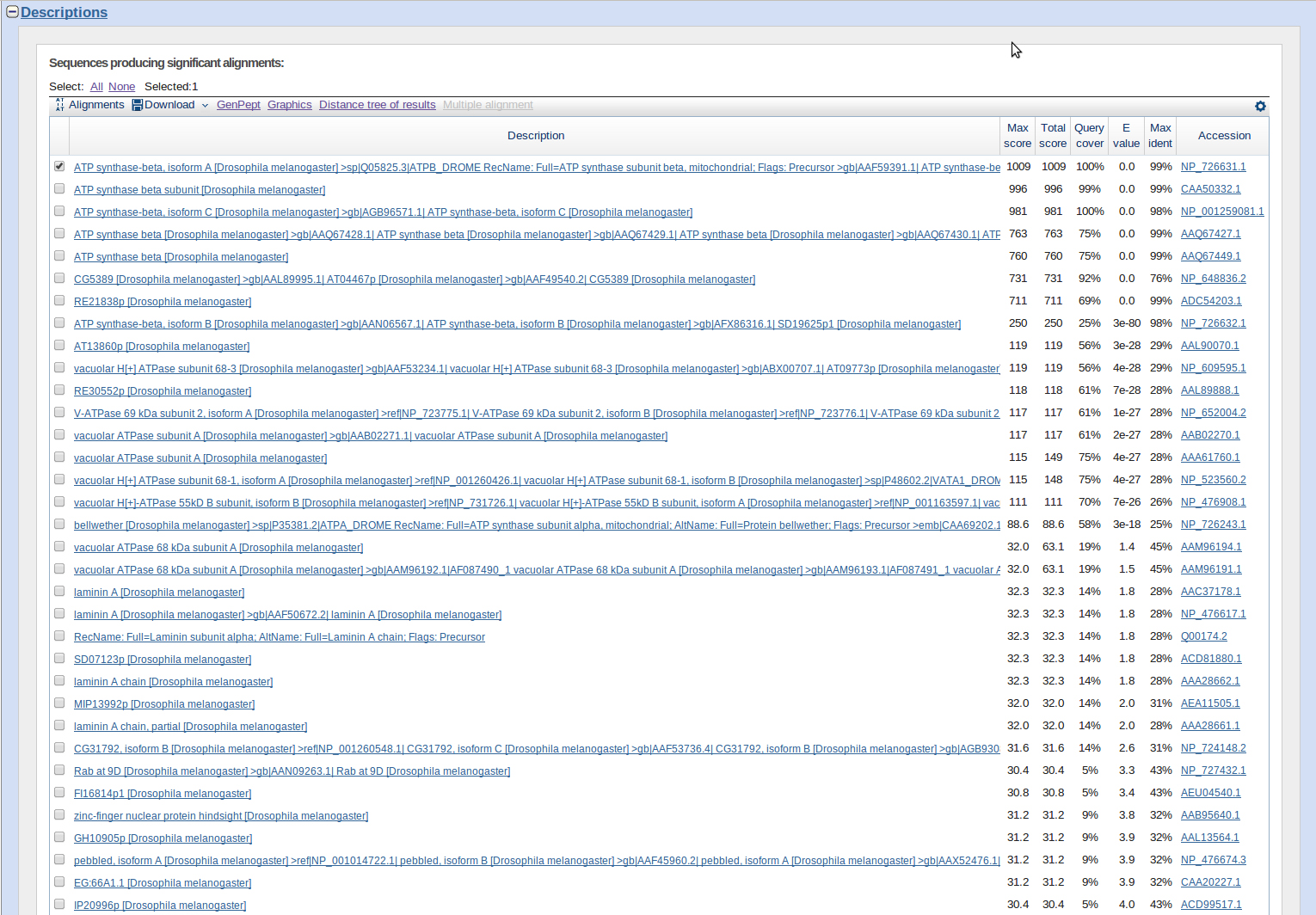
jub was a BLAST hit with good E-values and on chromosome 4. However it is out of order with the other three genes, ATPsyn-β, CaMKII, and Zyx. It appears to be further upstream.
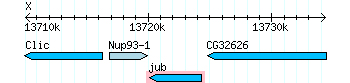
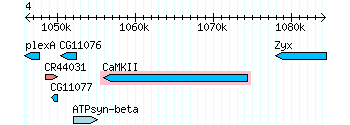
CaMKI resulted in a hit because since it is in the same family as CaMKII there are some shared sequences. If you take a look at the UCSC genome browser only a portion is shared with CaMKII; ~24500-25200 mp region.
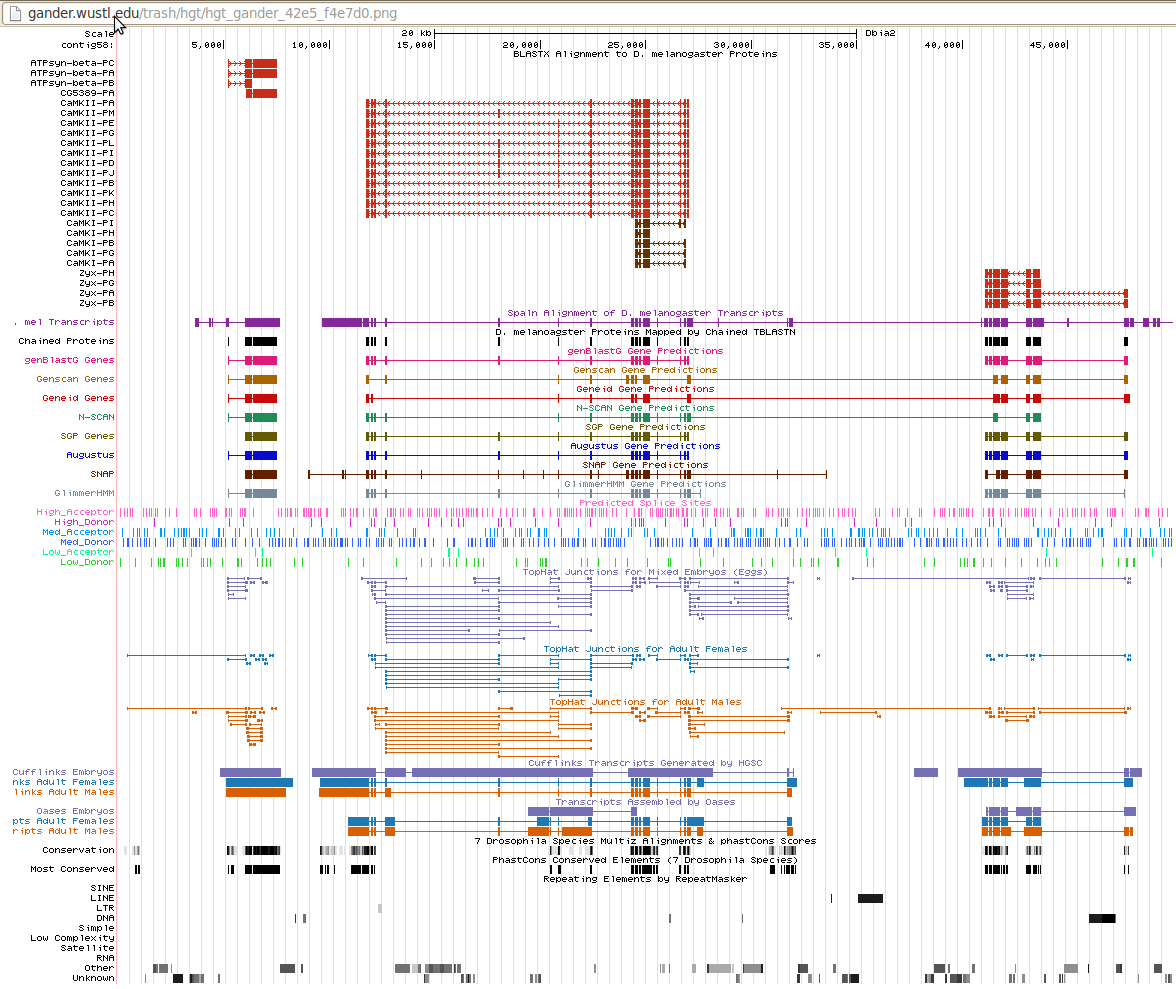
CG5389 came up on BLAST search as a result with good E-values. However its location on chromosome 3L concludes its insignificance in further examination. Our studies are concerned only with chromosome 4.
| ATPsyn-β | contig 58 | Alignment | |||||
| Start | End | Start | End | Frame | E | Identity | Positive |
| 1 | 21 | 5202 | 5264 | +3 | 0.017 | 100.00% | 100.00% |
| 55 | 132 | 6121 | 6354 | +1 | 0 | 100.00% | 100.00% |
| 131 | 511 | 6402 | 7544 | +3 | 0 | 95.50% | 96.10% |
| CaMKII | contig 58 | Alignment | |||||
| Start | End | Start | End | Frame | E | Identity | Positive |
| 7 | 22 | 27031 | 26984 | -2 | 3.75E-014 | 100.00% | 100.00% |
| 22 | 53 | 26910 | 26815 | -3 | 3.75E-014 | 96.90% | 100.00% |
| 54 | 73 | 26674 | 26614 | -2 | 0.002 | 100.00% | 100.00% |
| 74 | 92 | 25581 | 25581 | -3 | 0.017 | 100.00% | 100.00% |
| 93 | 201 | 25213 | 24887 | -2 | 1.26E-010 | 100.00% | 100.00% |
| 201 | 236 | 24801 | 24706 | -3 | 1.26E-144 | 100.00% | 100.00% |
| 233 | 273 | 24629 | 24507 | -1 | 1.26E-144 | 100.00% | 100.00% |
| 269 | 317 | 24460 | 24314 | -2 | 1.26E-144 | 98.90% | 95.90% |
| 347 | 365 | 20917 | 20861 | -2 | 0.715 | 89.50% | 94.70% |
| 387 | 413 | 12739 | 12659 | -2 | 9.64E-006 | 96.30% | 100.00% |
| 416 | 444 | 12236 | 12150 | -1 | 4.489E-055 | 100.00% | 100.00% |
| 444 | 530 | 12093 | 11773 | -3 | 4.48E-055 | 79.40% | 81.30% |
| Zyx | contig 58 | Alignment | |||||
| Start | End | Start | End | Frame | E | Identity | Positive |
| 1 | 65 | 47885 | 47688 | -1 | 1.49E-006 | 52.50% | 61.20% |
| 64 | 203 | 43756 | 43355 | -2 | 3.12E-053 | 66.40% | 78.60% |
| 210 | 287 | 43287 | 43048 | -3 | 3.12E-053 | 46.20% | 57.50% |
| 289 | 387 | 42159 | 41860 | -3 | 2.64E-001 | 48.50% | 63.40% |
| 386 | 502 | 41813 | 41463 | -1 | 2.64E-001 | 96.60% | 98.30% |
| 502 | 542 | 41406 | 41284 | -3 | 2.64E-001 | 92.70% | 97.60% |
| 543 | 585 | 41218 | 41090 | -2 | 2.64E-001 | 95.30% | 97.70% |