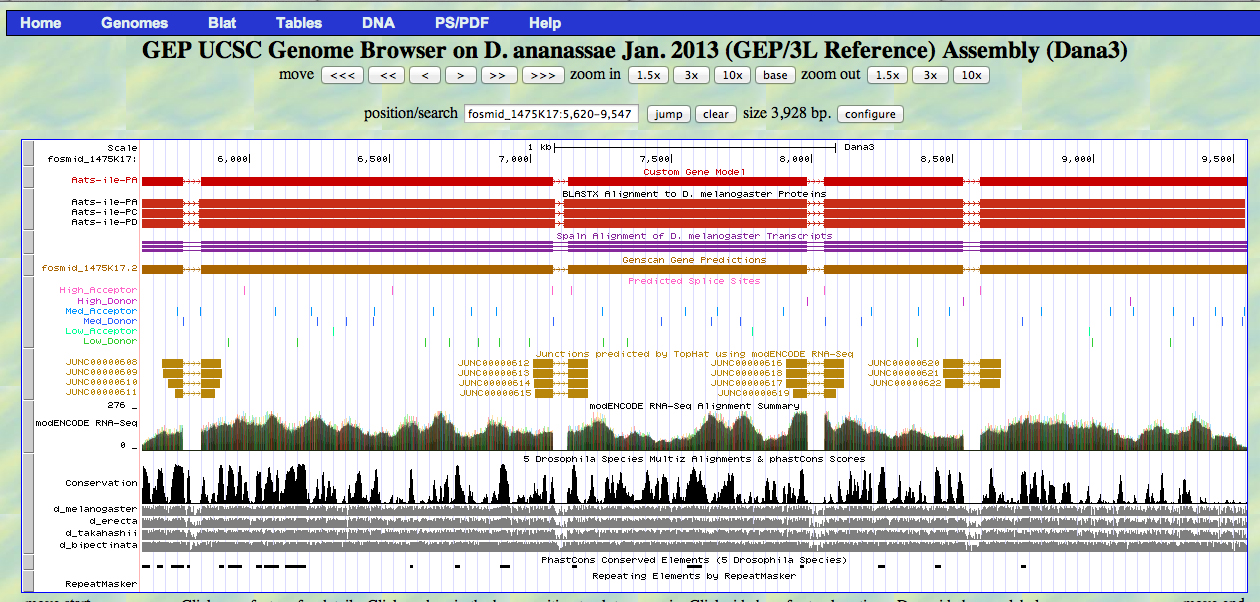
The D. ananassae Aats-ile gene is located on the plus strand of fosmid 1475K17.
The Gene Record Finder gives five CDS segments for D. melanogaster Aats-ile. There are three isoforms encoding identical proteins.
>Aats-ile:3_602_0 MGKKLERNDVCRVPENINFPAEEENVLQKWRHENIFEKCSQLSKGKP >Aats-ile:5_602_1 YTFYDGPPFATGLPHYGHILAGTIKDIVTRYAYQQGYHVDRRFGWDCHGL PVEFEIDKLLNIKGPEDVAKMGIAAYNAECRKIVMRYADEWENVVTRVGR WIDFKNDYKTLYPWYMESIWWIFKQLFDKGLVYQGVKVMPYSTACTTSLS NFEANQNYKEVVDPCVVVALEAVSLPNTFFLVWTTTPWTLPSNFACCVHP TMTYVKVRDVKSDRLFVLAESRLSYVYKSETEYEVKEKFVGKTLKDLHYK PLFPYFAKRGAEVKAYRVLVDEYVTEDSGTGIVHNAPYFGEDDYRVCLAA GLITKSSEVLCPVDEAGRFTNEASDFEGQYVKDSDKQIMAALKARGNLVS SGQVKHSYPFCWRSDTPLIYKAVPSWFVRVEHMSKNLLDCSSQTYWVPDF VKEKRFGNWLKEARDWA >Aats-ile:6_602_0 ISRNRYWGTPIPIWRSPSGDETVVIGSIKQLAELSGVQVEDLHRESIDHI EIPSAVPGNPPLRRIAPVFDCWFESGSMPFAQQHFPFENEKDFMNNFPAD FIAEGIDQTRGWFYTLLVISTALFNKAPFKNLIASGLVLAADGQKMSKRK KNYPDPMEVVHKYGADALRLYLINSPVVRAESLRFKEEGVRDIIKDVFLP WYNAYRFLLQNIVRYEKEDLAGNGQYTYDRERHLKNMDKASVIDVWILSF KESLLEFFATEMKMYRLYTVVPRLTKFIDQLTN >Aats-ile:7_602_1 YVRLNRRRIKGELGADQCIQSLDTLYDVLYTMVKMMAPFTPYLTEYIFQR LVLFQPAGTLEHADSVHYQMMPVSQKKFIRNDIERSVALMQSVVELGRVM RDRRTLPVKYPVSEIIAIHKDSQILEAIKTLQDFILSELNVRKLTLSSDK EKYGVTLRAEPDHK >Aats-ile:8_602_0 ALGQRLKGNFKAVMAAIKALRDDEIQKQVSQGYFDILDQRIELDEVRIIY CTSEQVGGNFEAHSDNEVLVLLDMTPNEELLEEGLAREVINRVQKLKKKA QLIPTDPVLIFHELAADNKAKQEVLEAQAQLAKVLSNYASIIKTAIKSEF APYSSEQASKKRLIASELVDLKGVPLKLTICSTEELQLPNLPWLNISLAE DLVPRFGNSSKASLFLQHNVSKEIISLPTLRSELEHLFGLYGVNFNIYVV DHQKRTTALKSIDENLSGKLLVLTRSQDAPKLSAGYELSPAPYSKFINQH SGKSIFTENPLGRALC*
I executed a bl2seq using BLASTX with the fosmid as the query sequence and the D. melanogaster CDS peptides above as subject sequences. Results are tabulated below.
| D. melanogaster Aats-ile CDS segments | fosmid | alignment | ||||
| start | end | frame | E | identity | positive | |
| Aats-ile:3_602_0 | 5621 | 5761 | +2 | 1e-25 | 89% | 97% |
| Aats-ile:5_602_1 | 5833 | 7080 | +1 | 0.0 | 96% | 98% |
| Aats-ile:6_602_0 | 7133 | 7981 | +2 | 0.0 | 97% | 98% |
| Aats-ile:7_602_1 | 8047 | 8538 | +1 | 2e-103 | 93% | 97% |
| Aats-ile:8_602_0 | 8600 | 9541 | +2 | 1e-161 | 77% | 87% |
The UCSC Genome Browser view below shows the 5' end of the first exon of D. ananassae Aats-ile.

In reading frame +2, there is an ATG codon (MET) beginning at 5621. This aligns to the D. melanogaster Aats-ile protein in the BLASTX track and to the GENSCAN model.
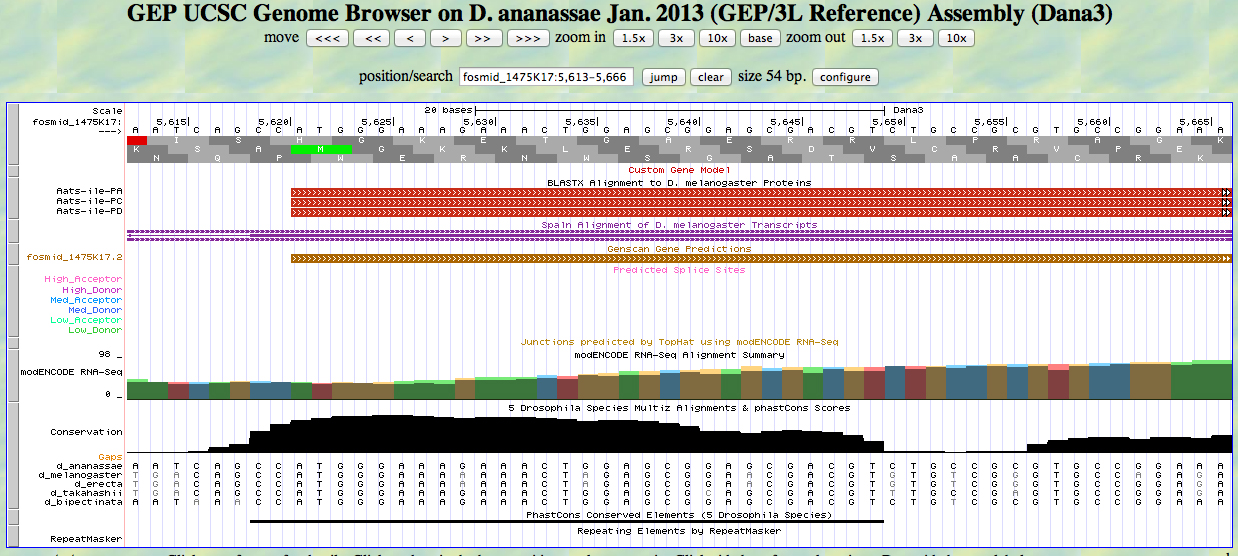
The UCSC Genome Browser view below shows the 3' end of the first exon of D. ananassae Aats-ile.
There is a GT at a medium donor site at the end of the GENSCAN model for the exon, near the end of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The last base of the coding sequence of the first exon is 5763.
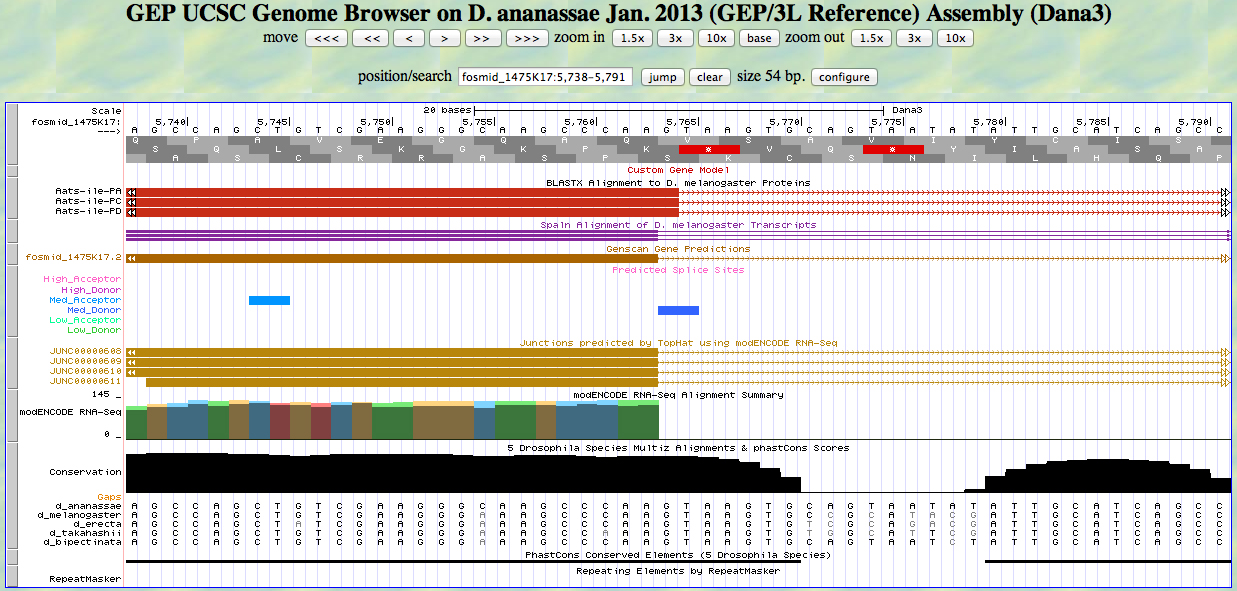
The UCSC Genome Browser view below shows the 5' end of the second exon of D. ananassae Aats-ile.
There is an AG at a medium acceptor site at the beginning of the GENSCAN model for the exon, near the beginning of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The first base of the coding sequence of the second exon is 5829.
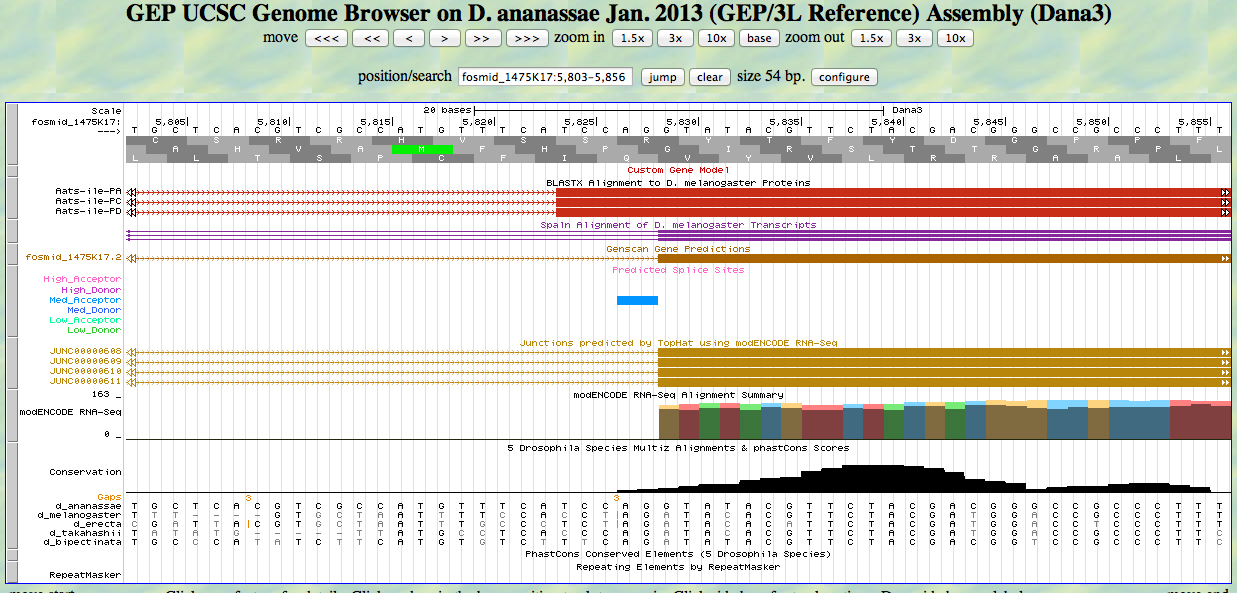
The UCSC Genome Browser view below shows the 3' end of the second exon and the 5' end of the third exon of D. ananassae Aats-ile.
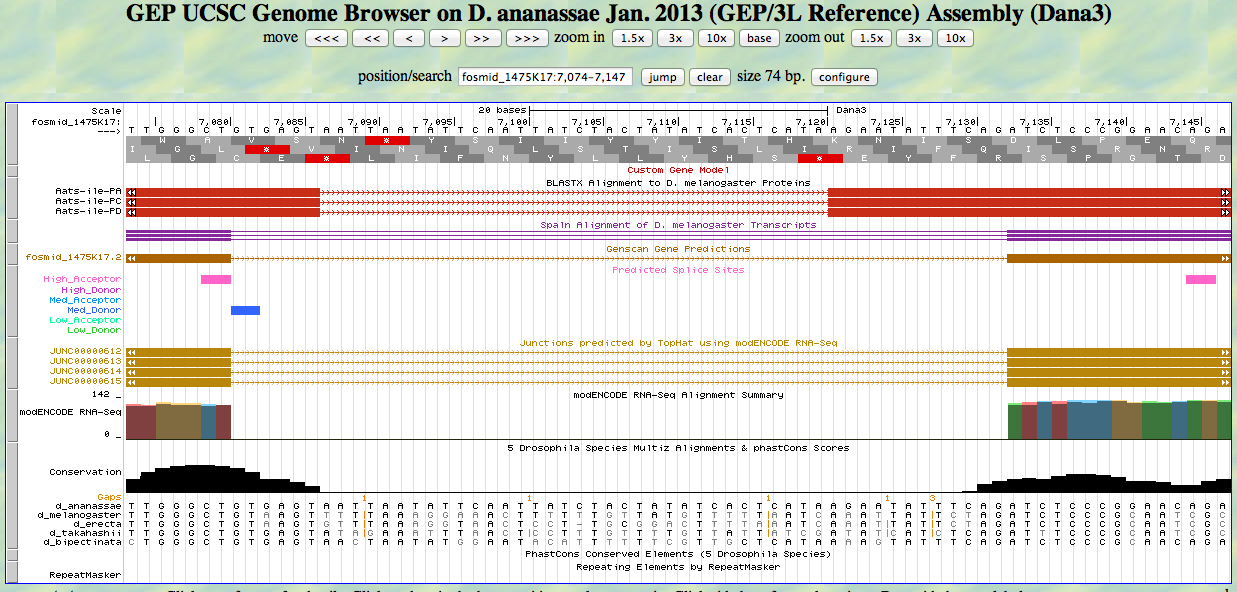
There is a GT at a medium donor site at the end of the GENSCAN model for the second exon, near the end of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The last base of the coding sequence of the second exon is 7080.
There is an AG at a unrated site at the beginning of the GENSCAN model for the third exon, near the beginning of the alignment to the D. melanogaster protein. The AG is at the position of a splice seen in the RNA-Seq (TopHat) data. The first base of the coding sequence of the third exon is 7133.
The UCSC Genome Browser view below shows the 3' end of the third exon and the 5' end of the fourth exon of D. ananassae Aats-ile.
There is a GT at a high donor site at the end of the GENSCAN model for the third exon, near the end of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The last base of the coding sequence of the third exon is 7983.
There is an AG at a high acceptor site at the beginning of the GENSCAN model for the fourth exon, near the beginning of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The first base of the coding sequence of the fourth exon is 8046.
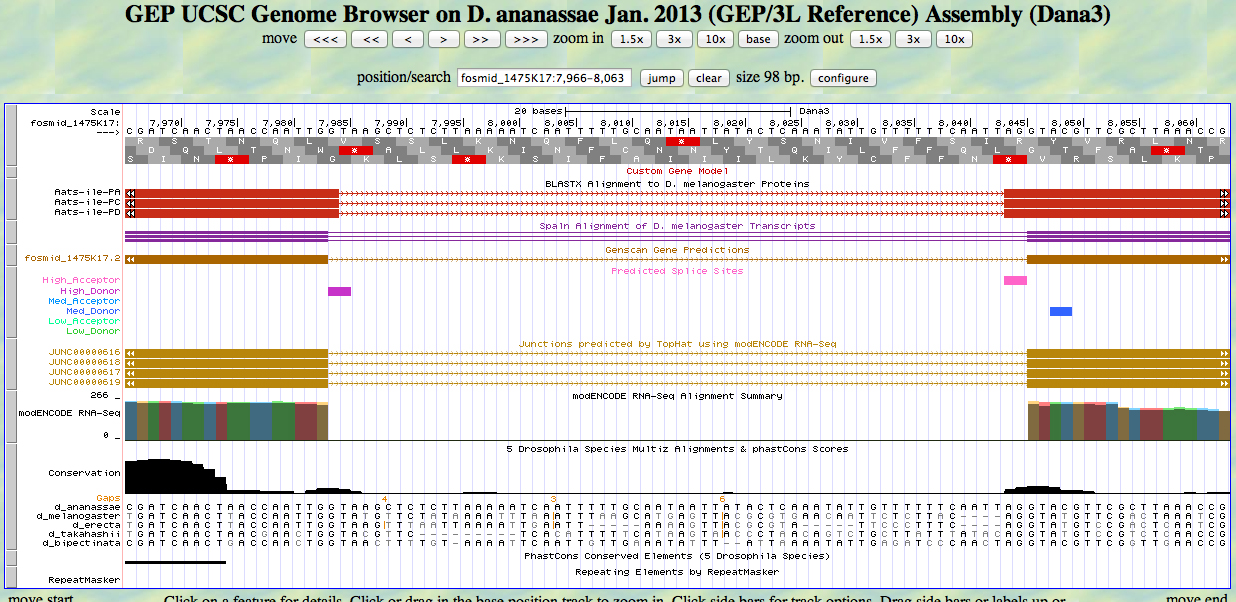
The UCSC Genome Browser view below shows the 3' end of the fourth exon and the 5' end of the fifth exon of D. ananassae Aats-ile.
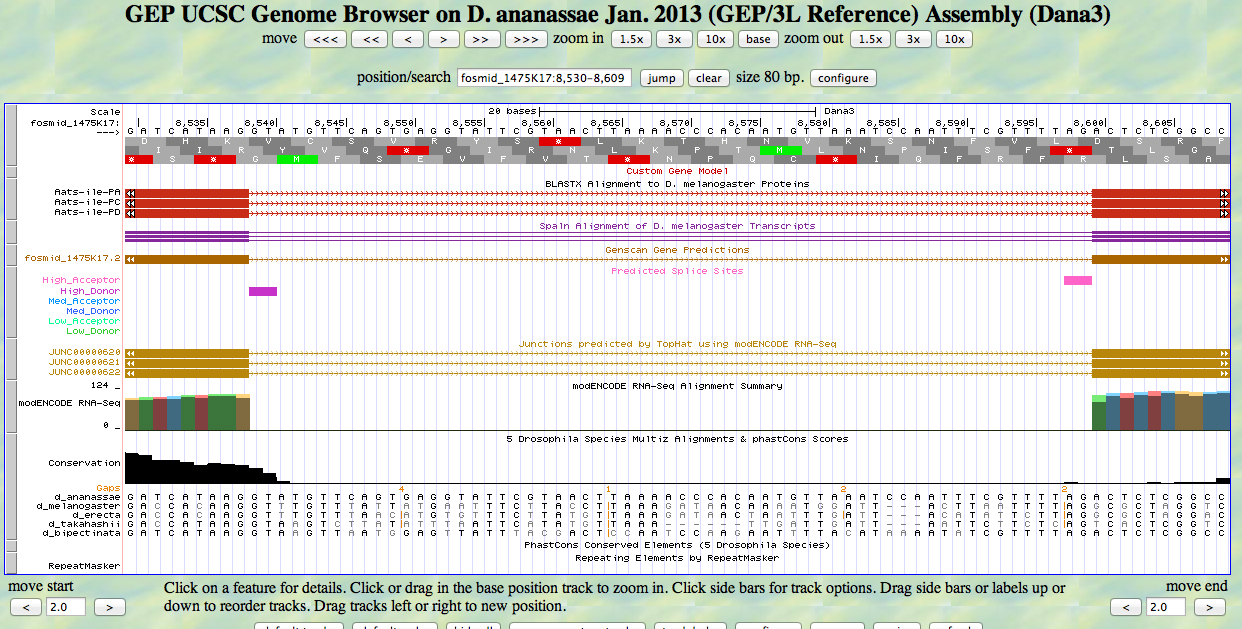
There is a GT at a high donor site at the end of the GENSCAN model for the fourth exon, at the end of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The last base of the coding sequence of the fourth exon is 8538.
There is an AG at a high acceptor site at the beginning of the GENSCAN model for the fifth exon, at the beginning of the alignment to the D. melanogaster protein. The predicted splice site matches the site of a splice seen in the RNA-Seq (TopHat) data. The first base of the coding sequence of the fifth exon is 8600.
The UCSC Genome Browser view below shows the 3' end of the fifth (last) exon of D. ananassae Aats-ile.
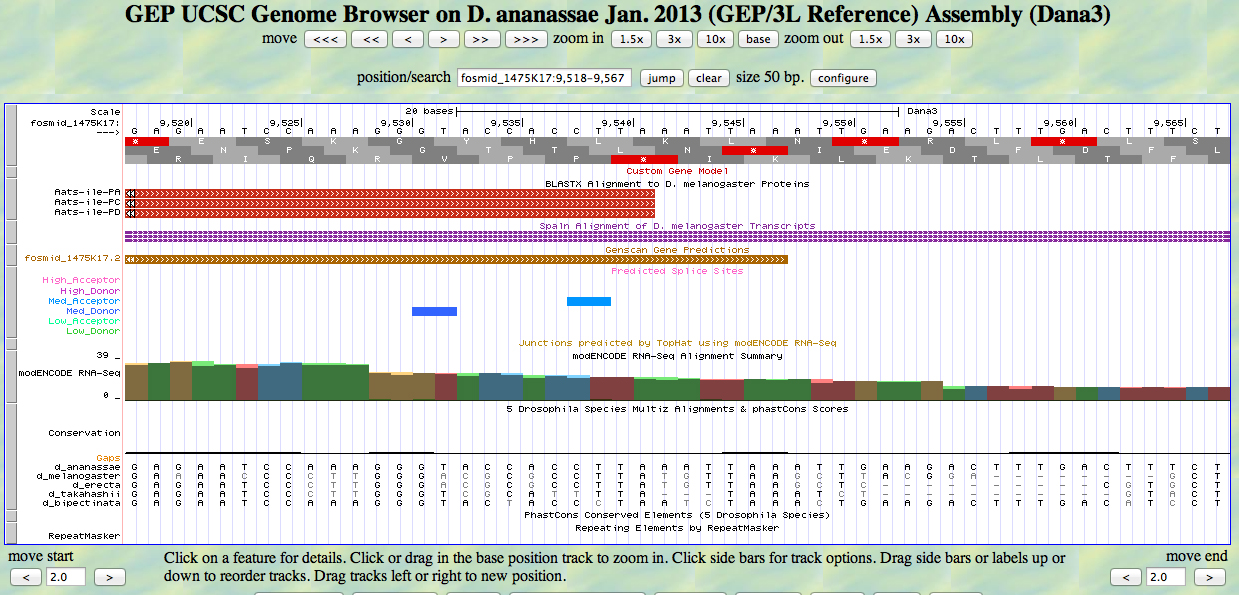
There is a TAA codon (STOP) in frame +2 at the end of the GENSCAN model for the exon, just past the end of the alignment to the D. melanogaster protein. The last base of the coding sequence of the fifth exon is 9544. The stop is at 9545-9547.
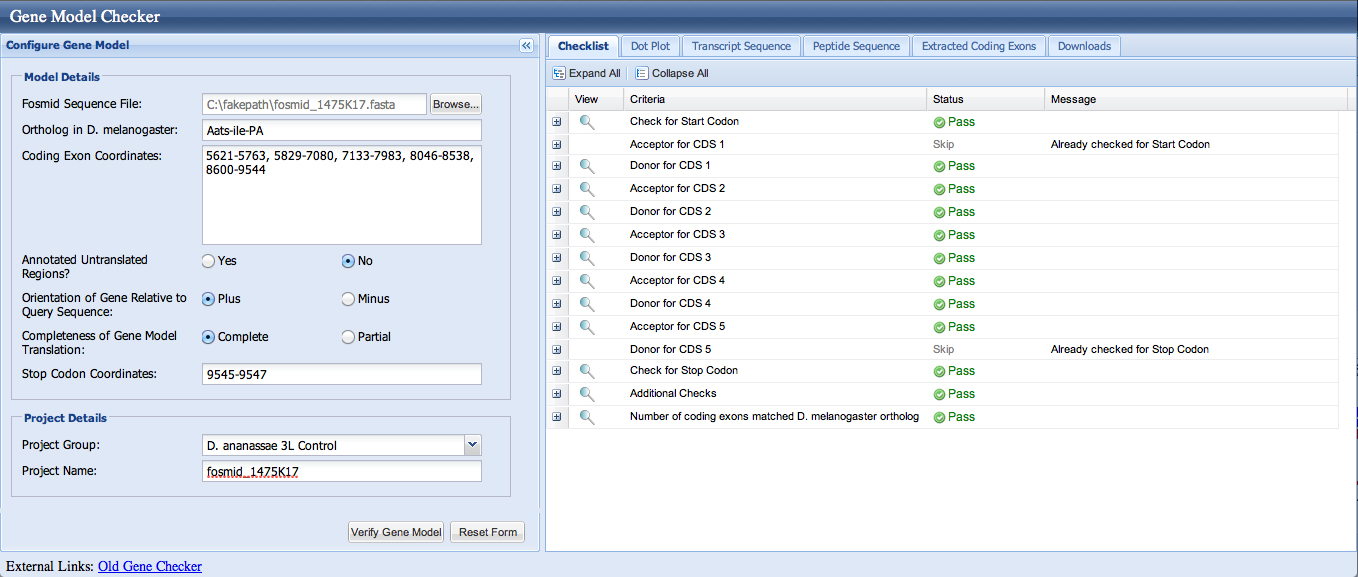
We enter the coordinates 5621-5763, 5829-7080, 7133-7983, 8046-8538, 8600-9544 into the Gene Model Checker; the checklist indicates no errors.
The dot matrix alignment of the D. melanogaster Aats-ile protein and the D. ananassae ortholog shows an excellent alignment.
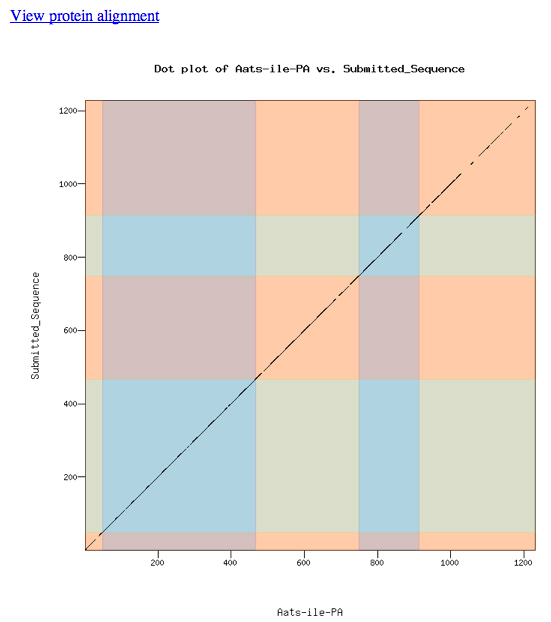
The alignment is excellent.

Loading the custom model into the UCSC Genome Browser produces the view shown below. The model aligns well to the D. melanogaster protein and the experimentally-observed splice sites.