The D. ananassae Hem gene is located on the minus strand of fosmid 1475K17.
The Gene Record Finder gives two CDS segments for D. melanogaster Hem:
>Hem:2_604_0 MARPIFPNQQKIAEKLIILNDRGLGILTRIYNIKKACGDTKSKPGFLSEK SLESSIKFIVKRFPNIDVKGLNAIVNIKAEIIKSLSLYYHTFVDLLDFKD NVCELLTTMDACQIHLDITLNFELTKYYLDLVVTYVSLMIVLSRVEDRKA VLGLYNAAYELQNNQADTGFPRLGQMILDYEVPLKKLAEEFIPHQRLLTS ALRSLTSIYALRNLPADKWREMQKLSLVGNPAILLKAVRTDTMSCEYISL EAMDRWIIFGLLLNHQMLGQYPEVNKIWLSALESSWVVALFRDEVLQIHQ YIQATFDGIKGYSKRIGEVKEAYNTAVQKAALMHRERRKFLRTALKELAL IMTDQPGLLGPKAIFIFIGLCLARDEILWLLRHNDNPPLLKNK >Hem:1_604_2 KSNEDLVDRQLPELLFHMEELRALVRKYSQVMQRYYVQYLSGFDATDLNI RMQSLQMCPEDESIIFSSLYNTAAALTVKQVEDNELFYFRPFRLDWFRLQ TYMSVGKAALRIAEHAELARLLDSMVFHTRVVDNLDEILVETSDLSIFCF YNKMFDDQFHMCLEFPAQNRYIIAFPLICSHFQNCTHEMCPEERHHIRER SLSVVNIFLEEMAKEAKNIITTICDEQCTMADALLPKHCAKILSVQSARK KKDKSKSKHFDDIRKPGDESYRKTREDLTTMDKLHMALTELCFAINYCPT VNVWEFAFAPREYLCQNLEHRFSRDLVGMVMFNQETMEIAKPSELLASVR AYMNVLQTVENYVHIDITRVFNNCLLQQTQALDSHGEKTIAALYNTWYSE VLLRRVSAGNIVFSINQKAFVPISPEGWVPFNPQEFSDLNELRALAELVG PYGIKTLNETLMWHIANQVQELKSLVSTNKEVLITLRTSFDKPEVMKEQF KRLQDVDRVLQRMTIIGVIICFRNLVHEALVDVLDKRIPFLLSSVKDFQE HLPGGDQIRVASEMASAAGLLCKVDPTLATTLKSKKPEFDEGEHLTACLL MVFVAVSIPKLARNENSFYRATIDGHSNNTHCMAAAINNIFGALFTICGQ SDMEDRMKEFLALASSSLLRLGQESDKEATRNRESIYLLLDEIVKQSPFL TMDLLESCFPYVLIRNAYHGVYKQEQILGLAL*
I executed a bl2seq using BLASTX with the fosmid as the query sequence and the D. melanogaster CDS peptides above as subject sequences. Results are tabulated below.
| D. melanogaster Hem CDS segments | fosmid | alignment | ||||
| start | end | frame | E | identity | positive | |
| Hem:2_604_0 | 4943 | 3765 | -1 | 0.0 | 97% | 98% |
| Hem:1_604_2 | 3700 | 1520 | -2 | 0.0 | 99% | 99% |
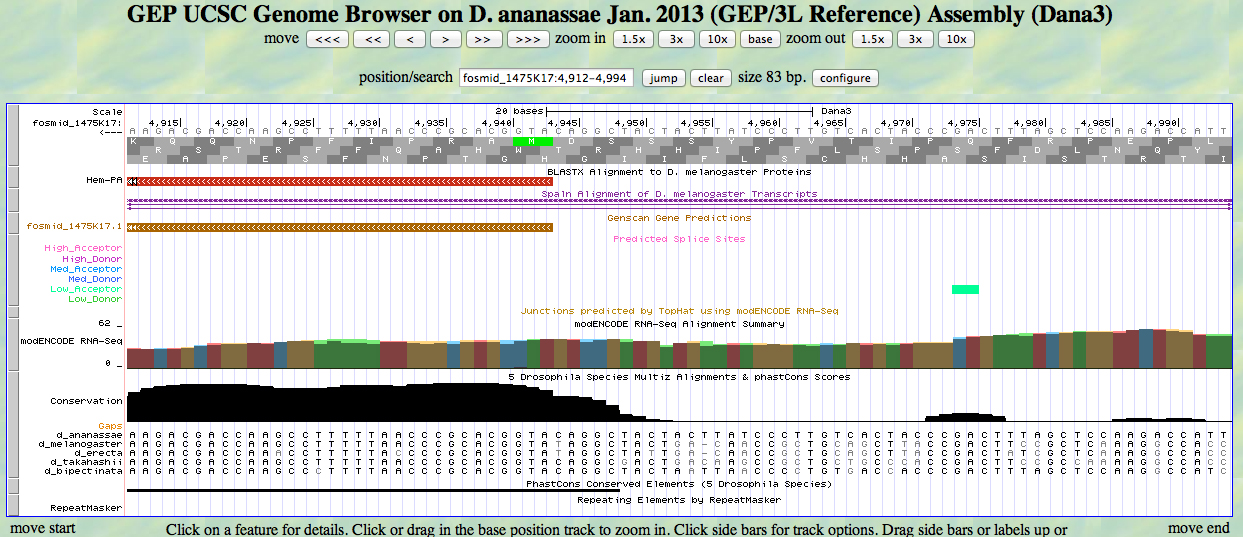
The UCSC Genome Browser view below shows the 5' end of D. ananassae Hem.

In reading frame -1, there is an ATG codon (MET) beginning at 4943. This aligns to the D. melanogaster Hem protein in the BLASTX track and to the GENSCAN model.
The UCSC Genome Browser view below shows the single intron in D. ananassae Hem.
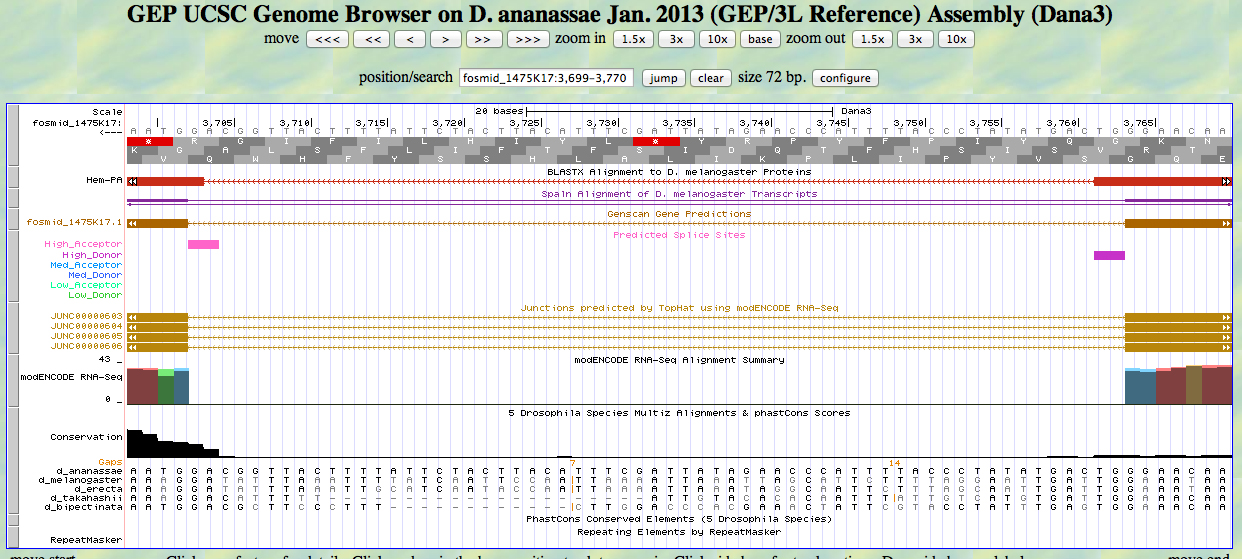
At the 5' end (right), we see the end of the alignment to the D. melanogaster Hem protein close to the position identified by RNA-Seq (TopHat) as a splice junction. There is a high donor splice site predicted at the GT. The last base of the first exon is 3764.
At the 3' end (left), we see the beginning of the alignment to the D. melanogaster Hem protein close to the position identified by RNA-Seq (TopHat) as a splice junction. There is a high acceptor splice site predicted at the AG. The first base of the second exon is 3702.
The UCSC Genome Browser view below shows the 3' end of the second exon of D. ananassae Hem.
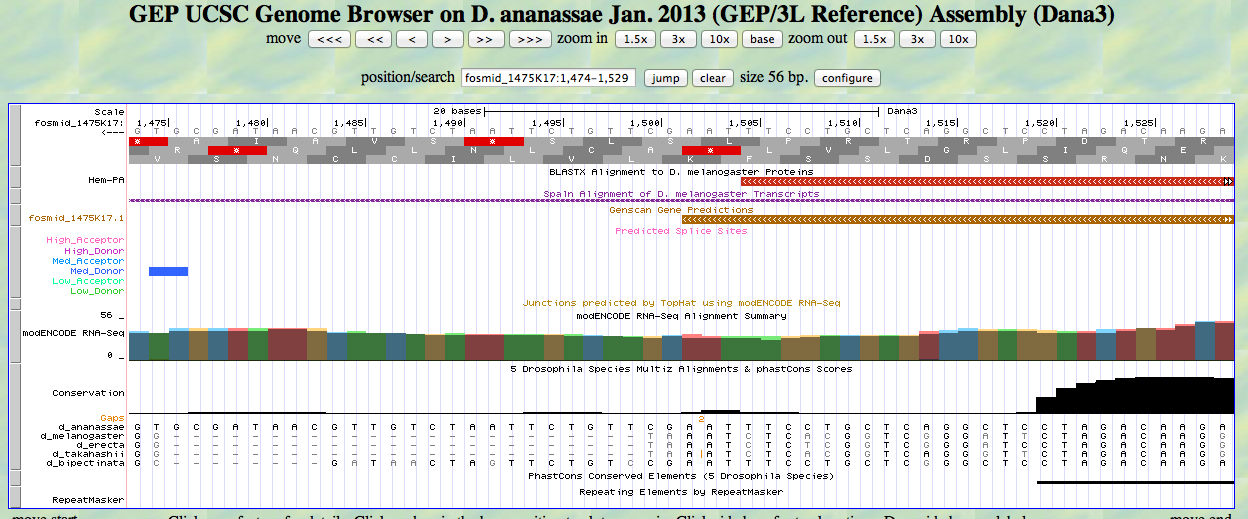
In reading frame -2, there is a TAA codon (STOP) at the end of the alignment to the D. melanogaster Hem protein. This is close to the end of the GENSCAN prediction. The last base of the second exon is 1505. The stop codon is 1504-1502.
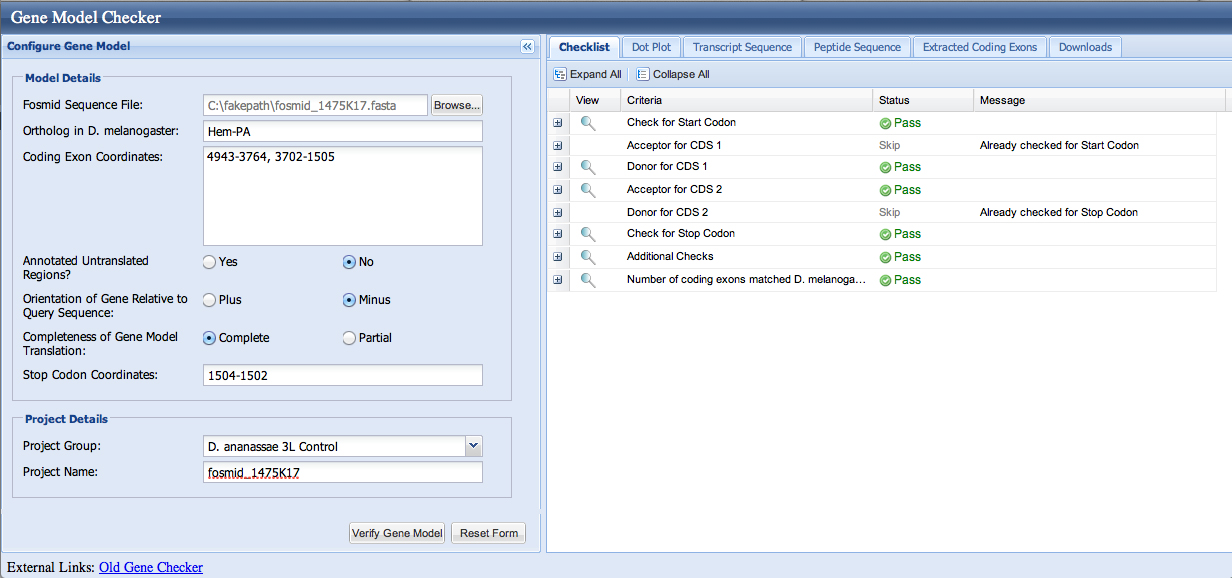
We enter the coordinates 4943-3764, 3702-1505 into the Gene Model Checker; the checklist indicates no errors.
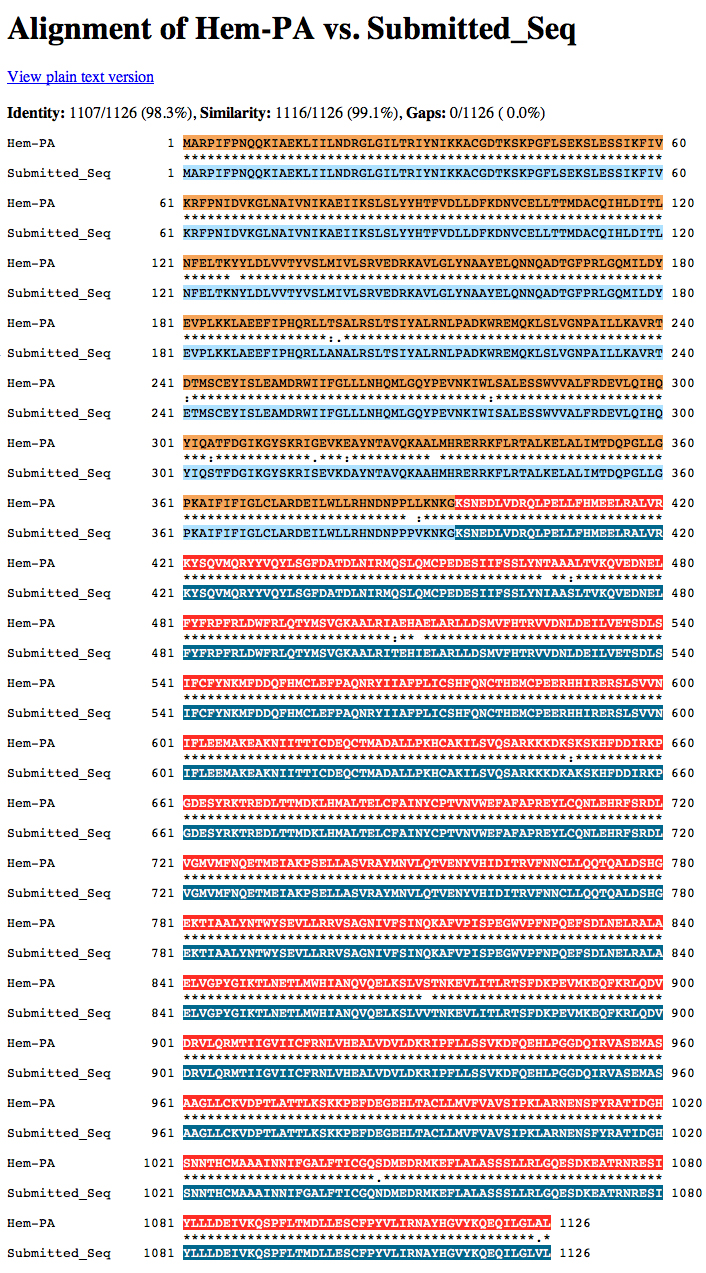
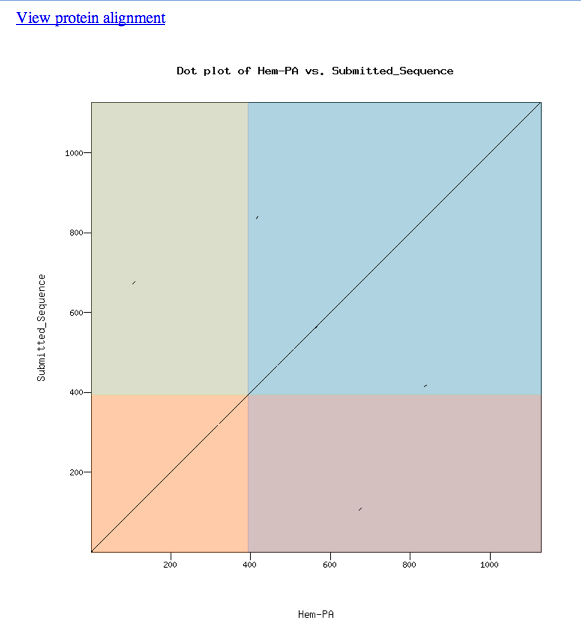
The dot matrix alignment of the D. melanogaster Hem protein and the D. ananassae ortholog shows an excellent alignment.
Loading the custom model into the UCSC Genome Browser produces the view shown below. The model aligns well to the D. melanogaster protein and the experimentally-observed splice sites.
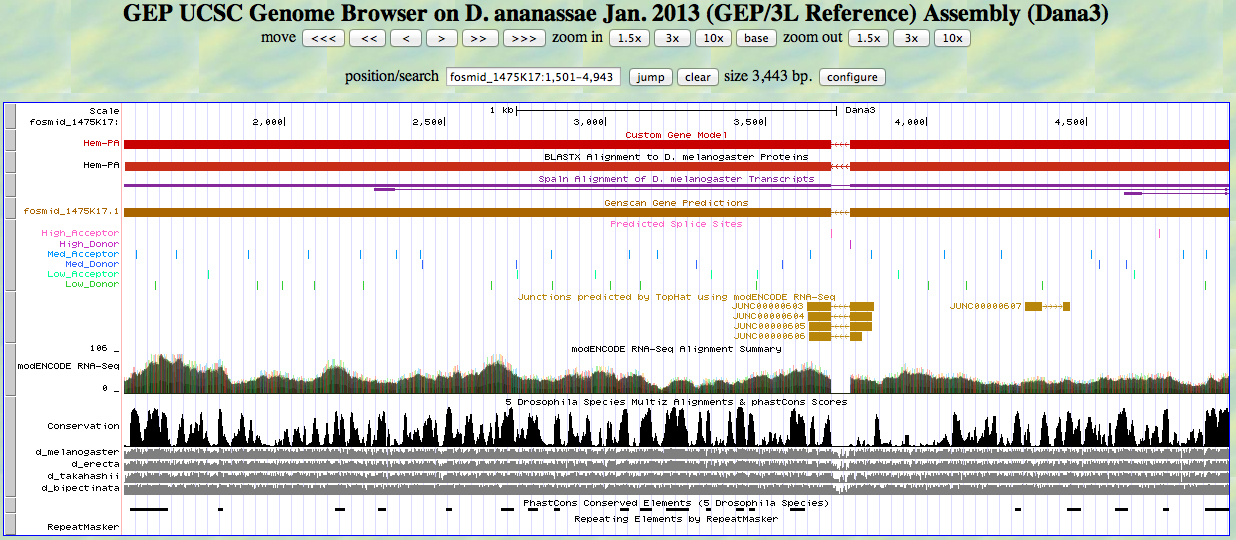
The alignment is excellent.