The D. ananassae Ten-m gene is located on the minus strand of fosmid 1475K17.
The Gene Record Finder gives thirteen CDS segments for D. melanogaster Ten-m:
>Ten-m:13_536_0 MNPYEYESTLDCRDVGGGPTPAHAHPHAQGRTLPMSGHGRPTTDLGPVHG SQTLQHQNQQNLQAAQAAAQSSHYDYEYQHLAHRPPDTANNTAQRTHGRQ >Ten-m:12_536_2 FLLEGVTPTAPPDVPPRNPTMSRMQNGRLTVNNPNDADFEPSCLVRTPSG NVYIPSGNL >Ten-m:11_536_2 INKGSPIDFKSGSACSTPTKDTLKGYERSTQGCMGPVLPQRSVMNGLPAH HYSAPMNFRKDLVARCSSPWFGIGSISVLFAFVVMLILLTTTGVIKWNQS PPCSVLVGNEASEVTAAKSTNTDLSKLHNSSVRAKNGQGIGLAQGQSGLG AAGVGSGGGSSAATVTTATSNSGTAQGLQSTSASAEATSSAATSSSQSSL TPSLSSSLANANNG >Ten-m:10_602_2 RAMSTRLSVRGAGERGRRHRRSLNEEQGEDDVATDGTFSDLITNESLNQQ AAEKYLATTLAKSPTDVHGSGNKTLPRMDGVYGTQRSEDTPDTSYDYVYE DEVEPETTPSLIRRTKTGQQFGKSLNSNLRSAAKTLVNKRRKYDHGTVEA EHIKHEEEEEEDEQKLERHEAIGMATELTTESETSTLPAVIDDDNQSDNS SSGPTPETTVRSDTDDIVEINTPPSQTAQRTFAAVSHQPAIEHDFQIKGT DAGGLQTEKPATDDINNERDLADNYEVDSKEPTSPGTPPQ >Ten-m:9_602_2 KVSQQTGKASLQSLQSESDLMMNDASHYEDIDIVKLDGLTISHEEEIYKT ADKENMAPKNQPSQHIDRSQNEVLKGHQQGDEKQPQLEPLKPYVSERVDL PGKRIFLNLTIATDEGSDSVYTLHVEVPTGGGPHFIKEVLTHEKPTAQAD SCVPEPPPRMPDCPCSCLPPPAPIYLDDTVDIDSAPPAKTVTTSTISAPI NPFHSEEEDDEDGVRDEEQTPSSSTATNLPSTEIDNHIAAFTEPAVGAGG VPFACPDVMPVLILE >Ten-m:8_536_2 ARTFPARSFPPDGTTFGQITLGQKLTKEIQPYSYWNMQFYQSEPAYVKFD YTIPRGASIGVYGRRNALPTHTQYHFKEVLSGFSASTRTARAAH >Ten-m:7_536_0 LSITREVTRYMEPGHWFVSLYNDDGDVQELTFYAAVAEDMTQNCPNGCSG NGQCLLGHCQCNPGFGGDDCSESVCPVLCSQHGEYTNGECICNPGWKGKE CSLRHDECEVADCSGHGHCVSGKCQCMRGYKGKFCEE >Ten-m:6_536_2 DCPHPNCSGHGFCADGTCICKKGWKGPDCATMDQDALQCLPDCSGHGTFD LDTQTCTCEAKWSGDDCSKELCDLDCGQHGRCEGDACACDPEWGGEYCNT RLCDVRCNEHGQCKNGTCLCVTGWNGKHCTIEGCPNSCAGHGQCRVSGEG QWECRCYEGWDGPDCGIALELNCGDSKDNDK >Ten-m:5_536_2 GLVDCEDPECCASHVCKTSQLCVSAPKPIDVLLRKQPPAITASFFERMKF LIDESSLQNYAKLETFNE >Ten-m:4_602_1 IFWNYFNA >Ten-m:3_536_1 RSAVIRGRVVTSLGMGLVGVRVSTTTLLEGFTLTRDDGWFDLMVNGGGAV TLQFGRAPFRPQSRIVQVPWNEVVIIDLVVMSMSEEKGLAVTTTHTCFAH DYDLMKPVVLASWKHGFQGACPDRSAILAESQVIQESLQIPGTGLNLVYH SSRAAGYLSTIKLQLTPDVIPTSLHLIHLRITIEGILFERIFEADPGIKF TYAWNRLNIYRQRVYGVTTAVVKVGYQYTDCTDIVWDIQTTKLSGHDMSI SEVGGWNLDIHHRYNFHEGILQKGDGSNIYLRNKPRIILTTMGDGHQRPL ECPDCDGQATKQRLLAPVALAAAPDGSLFVGDFNYIRRIMTDGSIRTVVK LNATRVSYRYHMALSPLDGTLYVSDPESHQIIRVRDTNDYSQPELNWEAV VGSGERCLPGDEAHCGDGALAKDAKLAYPKGIAISSDNILYFADGTNIRM VDRDGIVSTLIGNHMHKSHWKPIPCEGTLKLEEMHLRWPTELAVSPMDNT LHIIDDHMILRMTPDGRVRVISGRPLHCATASTAYDTDLATHATLVMPQS IAFGPLGELYVAESDSQRINRVRVIGTDGRIAPFAGAESKCNCLERGCDC FEAEHYLATSAKFNTIAALAVTPDSHVHIADQANYRIRSVMSSIPEASPS REYEIYAPDMQEIYIFNRFGQHVSTRNILTGETTYVFTYNVNTSNGKLST VTDAAGNKVFLLRDYTSQVNSIENTKGQKCRLRMTRMKMLHELSTPDNYN VTYEYHGPTGLLRTKLDSTGRSYVYNYDEFGRLTSAVTPTGRVIELSFDL SVKGAQVKVSENAQKEMSLLIQGATVIVRNGAAESRTTVDMDGSTTSITP WGHNLQMEVAPYTILAEQSPLLGESYPVPAKQRTEIAGDLANRFEWRYFV RRQQPLQAGKQSKGPPRPVTEVGRKLRVNGDNVLTLEYDRETQSVVVMVD DKQELLNVTYDRTSRPISFRPQSGDYADVDLEYDRFGRLVSWKWGVLQEA YSFDRNGRLNEIKYGDGSTMVYAFKDMFGSLPLKVTTPRRSDYLLQYDDA GALQSLTTPRGHIHAFSLQTSLGFFKYQYYSPINRHPFEILYNDEGQILA KIHPHQSGK >Ten-m:1_536_0 VAFVHDTAGRLETILAGLSSTHYTYQDTTSLVKSVEVQEPGFELRREFKY HAGILKDEKLRFGSKNSLASARYKYAYDGNARLSGIEMAIDDKELPTTRY KYSQNLGQLEVVQDLKITRNAFNRTVIQDSAKQFFAIVDYDQHGRVKSVL MNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNADGHVVEVLG TNNWKYLFDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGF VVKRGEQKYRYNNRGQLIHSFERERFQSWYYYDDRSRLVAWHDNKGNTTQ YYYANPRTPHLVTHVHFPKISRTMKLFYDDRDMLIALEHEDQRYYVATDQ NGSPLAFFDQNGSIVKEMKRTPFGRIIKDTKPEFFVPIDFHGGLIDPHTK LVYTEQRQYDPHVGQWMTPLWETLATEMSHPTDVFIYRYHNNDPINPNKP QNYMIDLDSWLQLFGYDLNNMQSSRYTKLAQYTPQASIKSNTLAPDFGVI SGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRVSYRRGVFGEGVL LSRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVK DNVLKLRDDNEELRRLGGMFNISTHEISDHGGSAAKELRLHGPDAVVIIK YGVDPEQERHRILKHAHKRAVERAWELEKQLVAAGFQGRGDWTEEEKEEL VQHGDVDGWNGIDIHSIHKYPQLADDPGNVAFQRDAKRKRRKTGSSHRSA SNRRQLKFGELSA* >Ten-m:2_602_0 VAFVHDTAGRLETILAGLSSTHYTYQDTTSLVKSVEVQEPGFELRREFKY HAGILKDEKLRFGSKNSLASARYKYAYDGNARLSGIEMAIDDKELPTTRY KYSQNLGQLEVVQDLKITRNAFNRTVIQDSAKQFFAIVDYDQHGRVKSVL MNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNADGHVVEVLG TNNWKYLFDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGF VVKRGEQKYRYNNRGQLIHSFERERFQSWYYYDDRSRLVAWHDNKGNTTQ YYYANPRTPHLVTHVHFPKISRTMKLFYDDRDMLIALEHEDQRYYVATDQ NGSPLAFFDQNGSIVKEMKRTPFGRIIKDTKPEFFVPIDFHGGLIDPHTK LVYTEQRQYDPHVGQWMTPLWETLATEMSHPTDVFIYRYHNNDPINPNKP QNYMIDLDSWLQLFGYDLNNMQSSRYTKLAQYTPQASIKSNTLAPDFGVI SGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRVSYRRGVFGEGVL LSRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVK DNVLKLRDDNEELRRLGGMFNISTHEISDHGGSAAKELRLHGPDAVVIIK YGVDPEQERHRILKHAHKRAVERAWELEKQLVAAGFQGRGDWTEEEKEEL VQHGDVDGWNGIDIHSIHKYPQLADDPGNVAFQRDAKRKRRKTGSSHRSA SNRRQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGK MEPRDSSEALDEQIL*
I executed a bl2seq using BLASTX with the fosmid as the query sequence and the D. melanogaster CDS peptides above as subject sequences. Results are tabulated below. We already know that CDS 12 and CDS 13 are not on the fosmid, so they are omitted from the table.
| D. melanogaster Ten-m CDS segments | BLASTX | |||||
| fosmid | alignment | |||||
| start | end | frame | E | identity | positive | |
| Ten-m:11_536_2 | 25756 | 25316 | -2 | 7e-66 | 80% | 86% |
| Ten-m:10_602_2 | 24496 | 23612 | -2 | 2e-54 | 48% | 62% |
| Ten-m:9_602_2 | 23192 | 22365 | -1 | 6e-68 | 59% | 68% |
| Ten-m:8_536_2 | 21938 | 21657 | -1 | 1e-60 | 99% | 100% |
| Ten-m:7_536_0 | 21528 | 21118 | -3 | 2e-92 | 98% | 99% |
| Ten-m:6_536_2 | 21025 | 20483 | -2 | 2e-114 | 99% | 99% |
| Ten-m:5_536_2 | 20421 | 20218 | -3 | 1e-42 | 100% | 100% |
| Ten-m:4_602_1 | 19180 | 19157 | -2 | 0.036 | 100% | 100% |
| Ten-m:3_536_1 | 18563 | 15240 | -1 | 0.0 | 98% | 99% |
| Ten-m:1_536_0 | 15175 | 12884 | -2 | 0.0 | 96% | 98% |
| Ten-m:2_602_0 | 15175 | 12701 | -2 | 0.0 | 92% | 95% |
The alignment of D. melanogaster CDS 4 to the fosmid is of marginal significance because the sequences is so short.
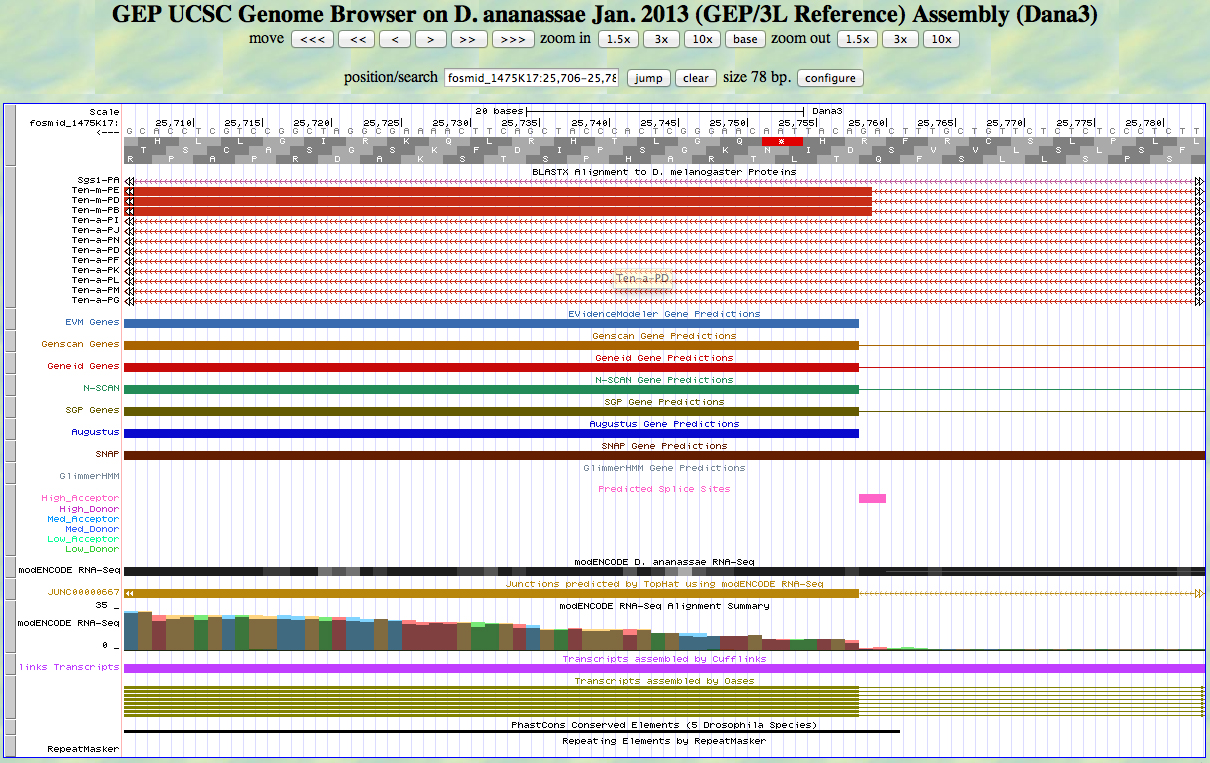
We begin with CDS 11. Here is the 5' end of the segment aligning to CDS 11 in the UCSC Genome Browser.

There is a high acceptor site predicted at the AG at the position of the alignment to the D. melanogaster protein, matching the 5' end predicted by most gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 25,758.
Because we are in frame -2, the phase of the exon is 0.
Here is the 3' end of the segment aligning to CDS 11 in the UCSC Genome Browser.
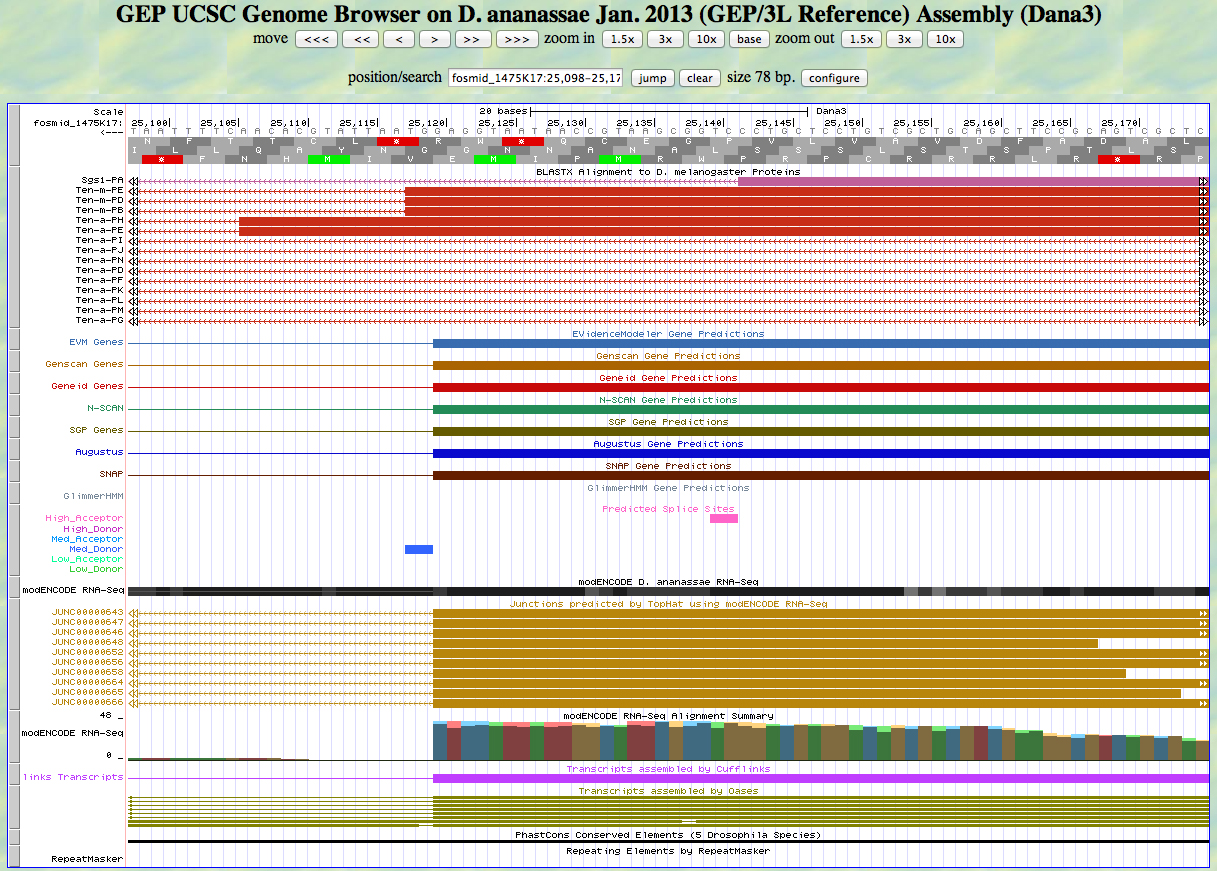
There is a medium donor predicted at the GT at the position of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 25,120.
Because we are in frame -2, the phase of the next exon is 0.
Here is the 5' end of the segment aligning to CDS 10 in the UCSC Genome Browser.

There is an AG predicted to be a medium acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 24,498.
Because we are in frame -2, the phase of the exon is 0.
Here is the 3' end of the segment aligning to CDS 10 in the UCSC Genome Browser.
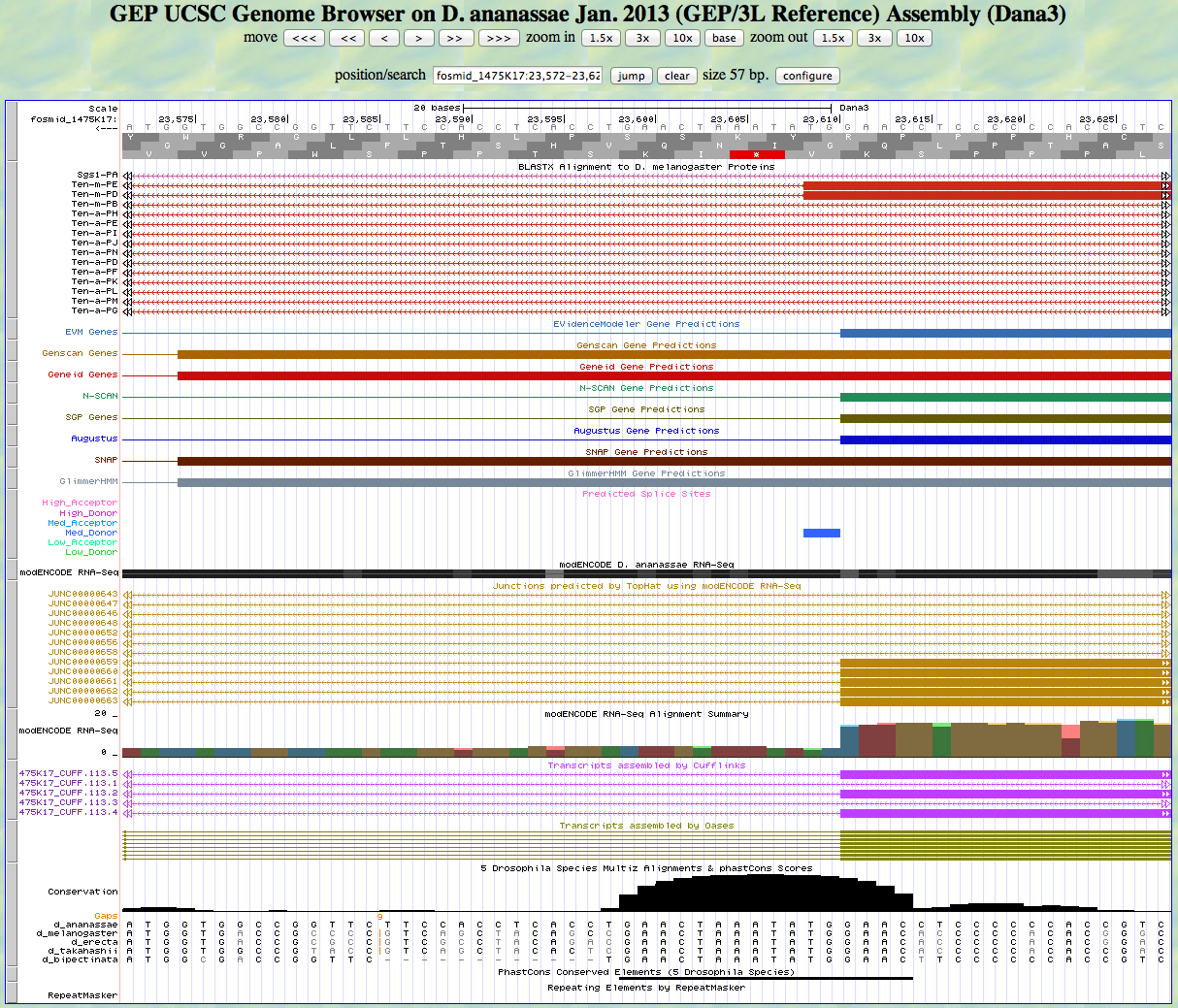
There is a GT predicted to be a medium donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by most gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 23,611.
Because we are in frame -2, the phase of the next exon is 0.
Here is the 5' end of the segment aligning to CDS 9 in the UCSC Genome Browser.

There are two possible splice acceptors here. One is an AG not identified as an acceptor at 23,250. There are four TopHat junctions here, and a strong species conservation signal at this position. The first base of the exon given this splice is 23,248.
The other is an AG identified as a medium acceptor at 23,202. There is a single TopHat junction here and a weaker conservation signal. The first base of the exon given this splice is 23,200.
The frame for this exon is -1. The previous exon, which we predict with confidence, gives the phase of this exon as 0. If the first base is 23,248, the phase is 0. If the first base is 23,200, the phase is also 0.
Here is the 3' end of the segment aligning to CDS 9 in the UCSC Genome Browser.
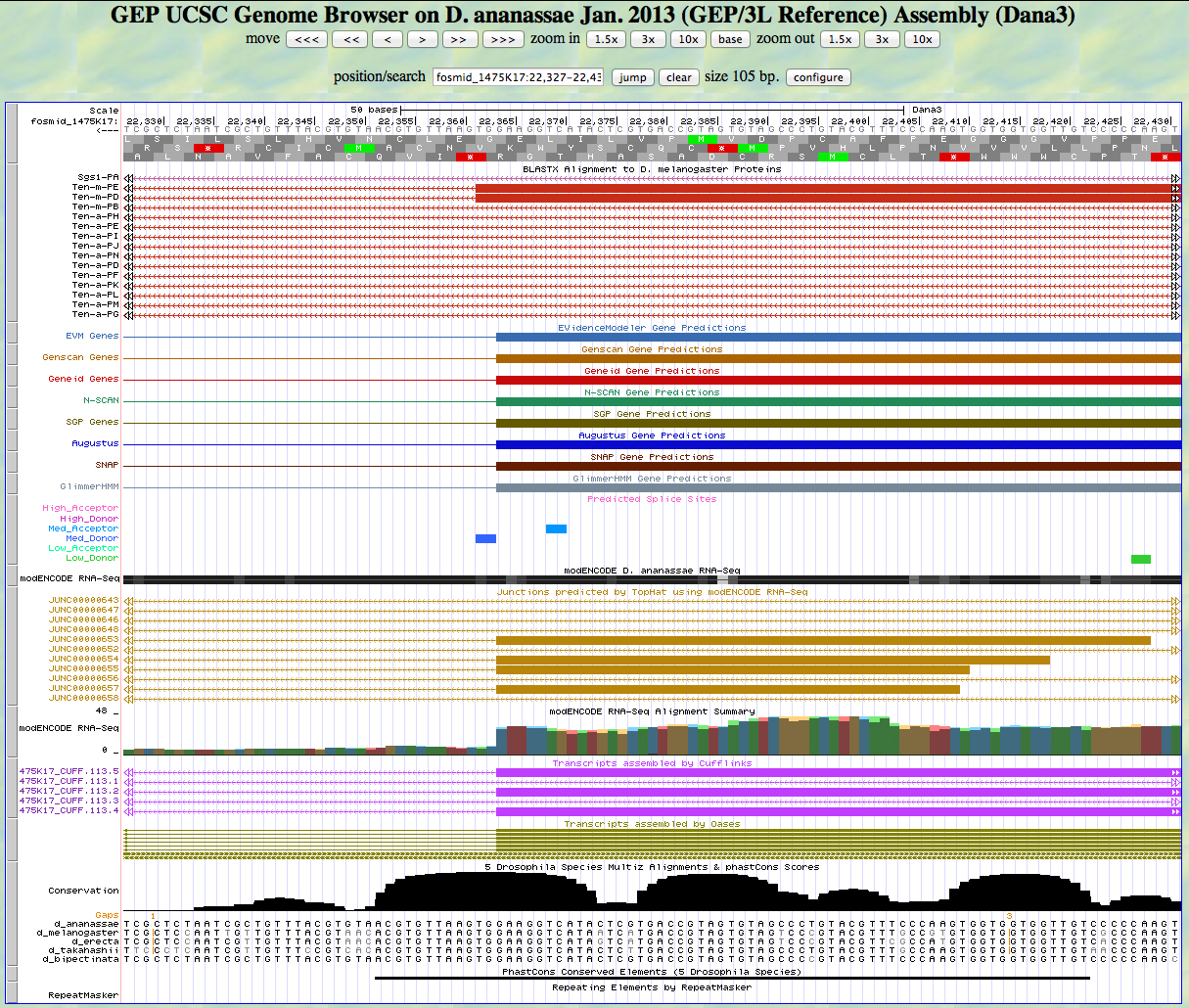
There is a GT predicted to be a medium donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 22,364.
Because we are in frame -1, the phase of the next exon is 2.
Here is the 5' end of the segment aligning to CDS 8 in the UCSC Genome Browser.
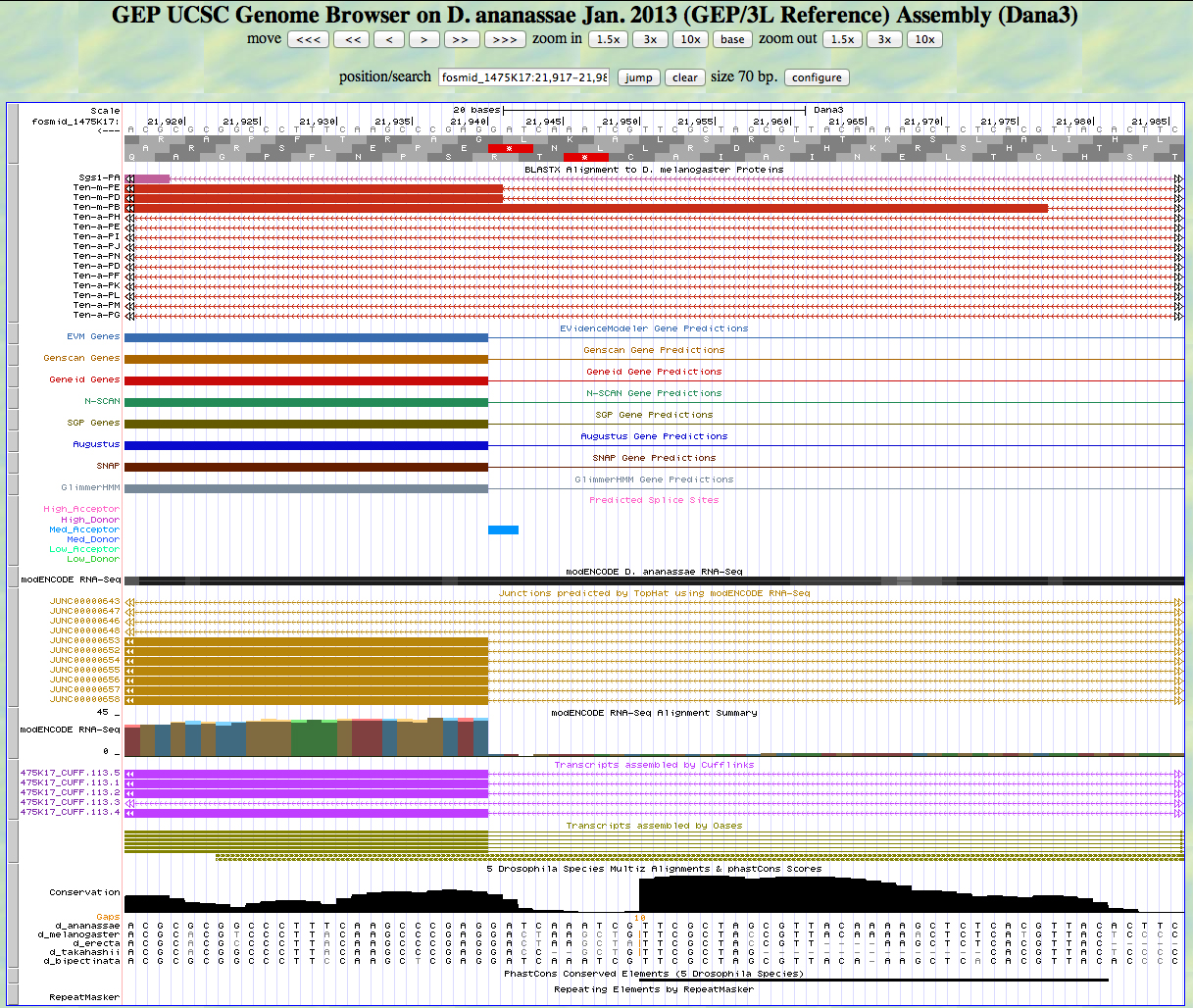
There is an AG predicted to be a medium acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 21,940.
Here is the 3' end of the segment aligning to CDS 8 in the UCSC Genome Browser.
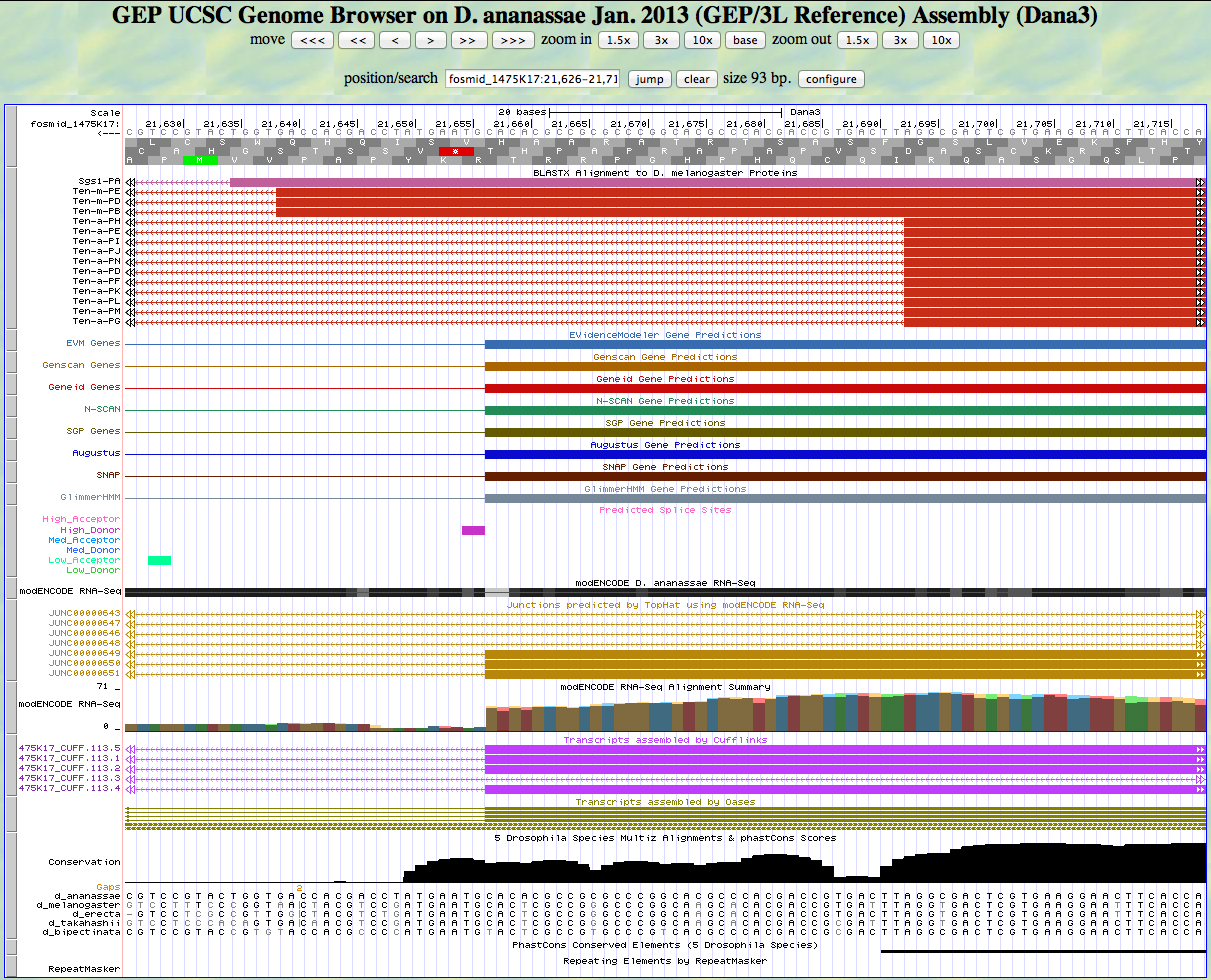
There is a GT predicted to be a high donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 21,657.
Here is the 5' end of the segment aligning to CDS 7 in the UCSC Genome Browser.
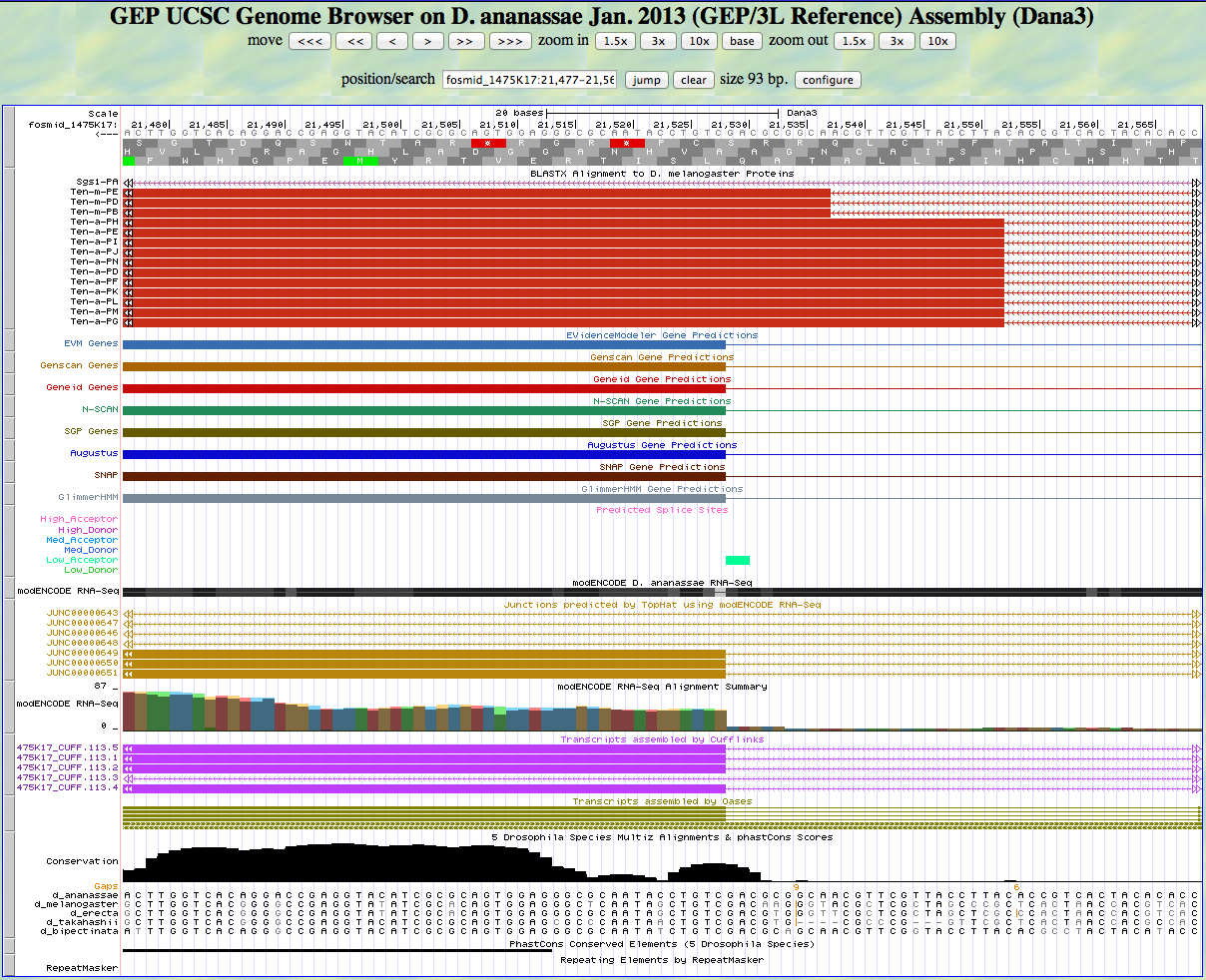
There is an AG predicted to be a low acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 21,528.
Here is the 3' end of the segment aligning to CDS 7 in the UCSC Genome Browser.
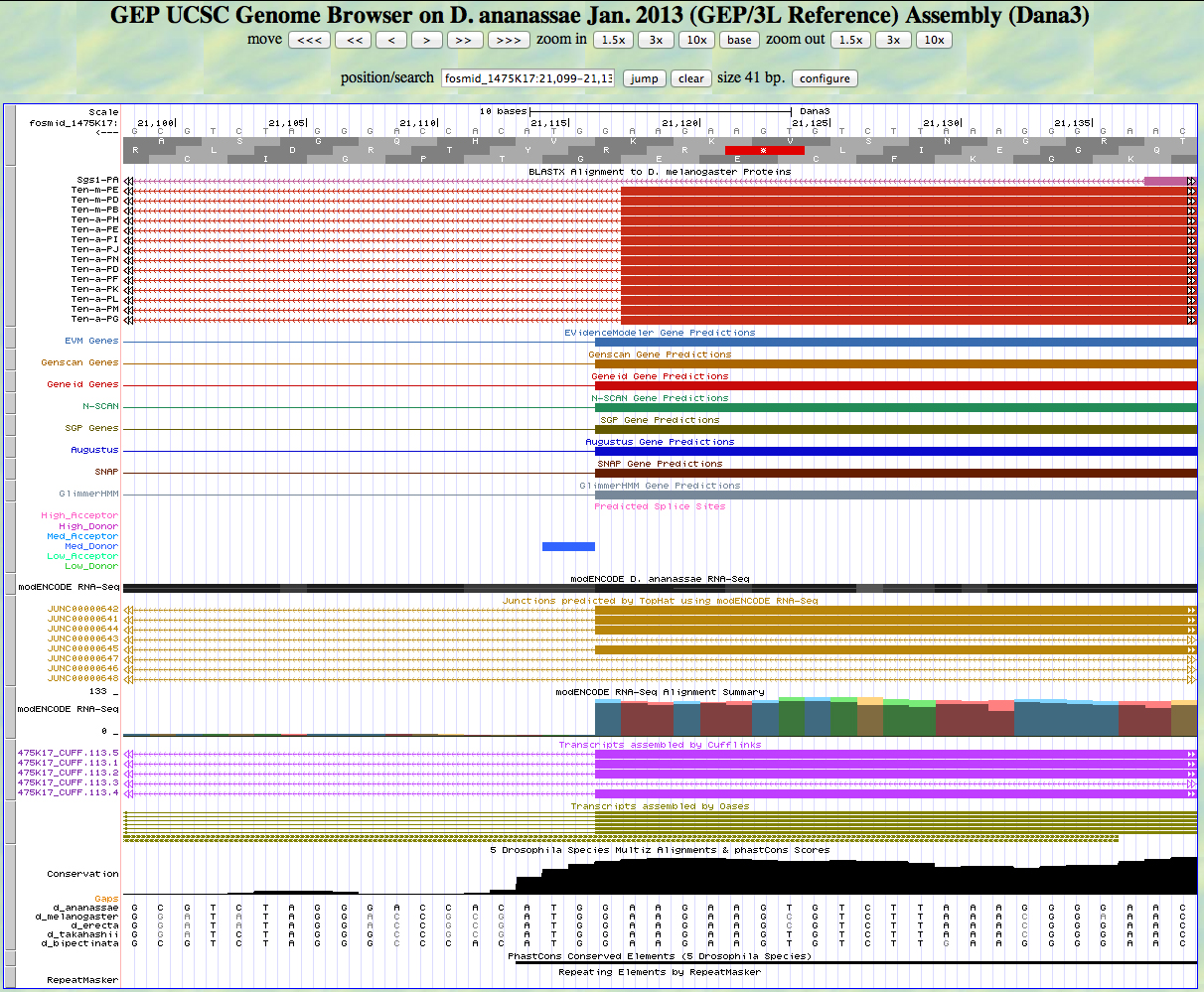
There is a GT predicted to be a medium donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well.The last base of this exon is 21,117.
Here is the 5' end of the segment aligning to CDS 6 in the UCSC Genome Browser.
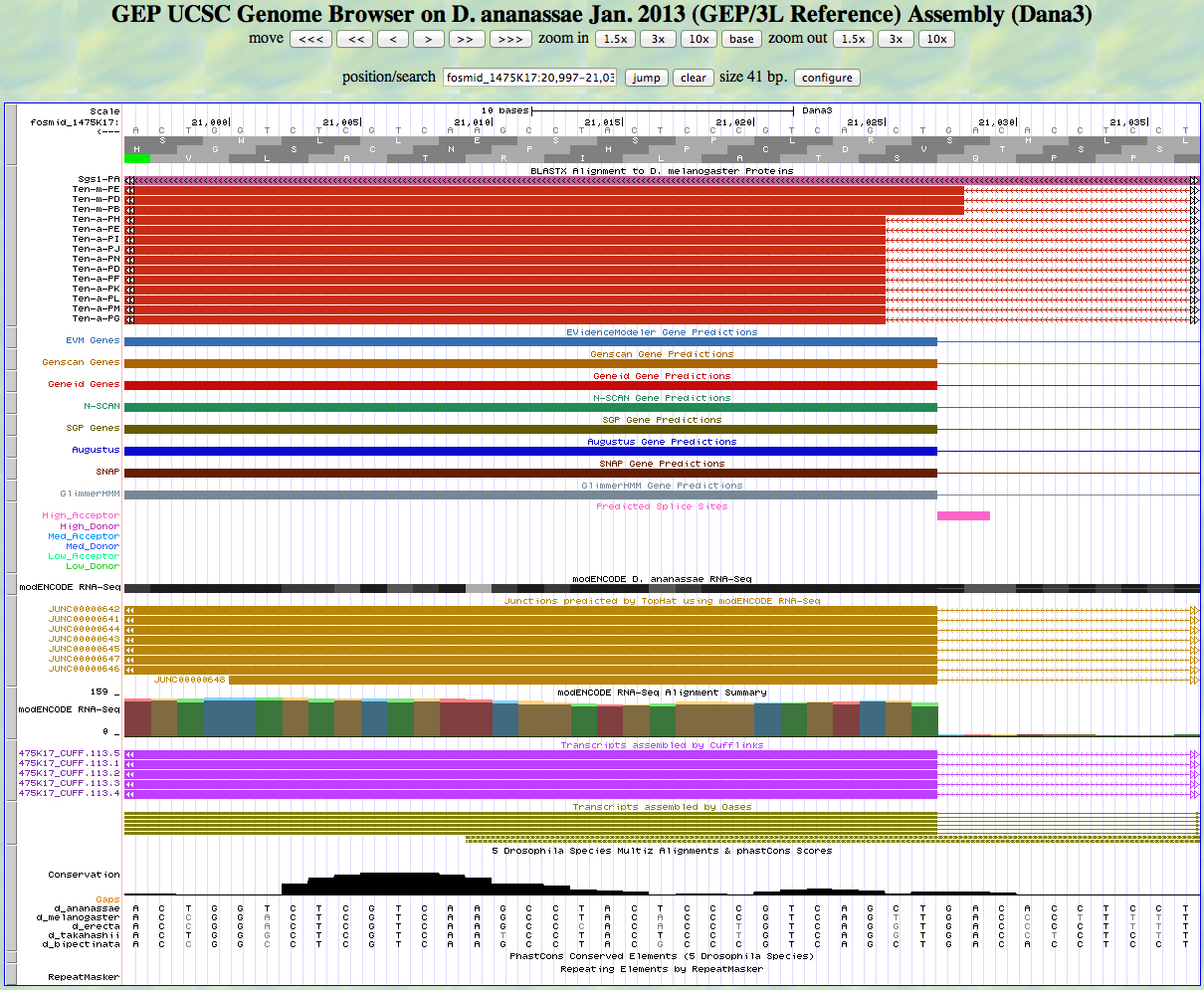
There is an AG predicted to be a high acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 21,027.
Here is the 3' end of the segment aligning to CDS 6 in the UCSC Genome Browser.
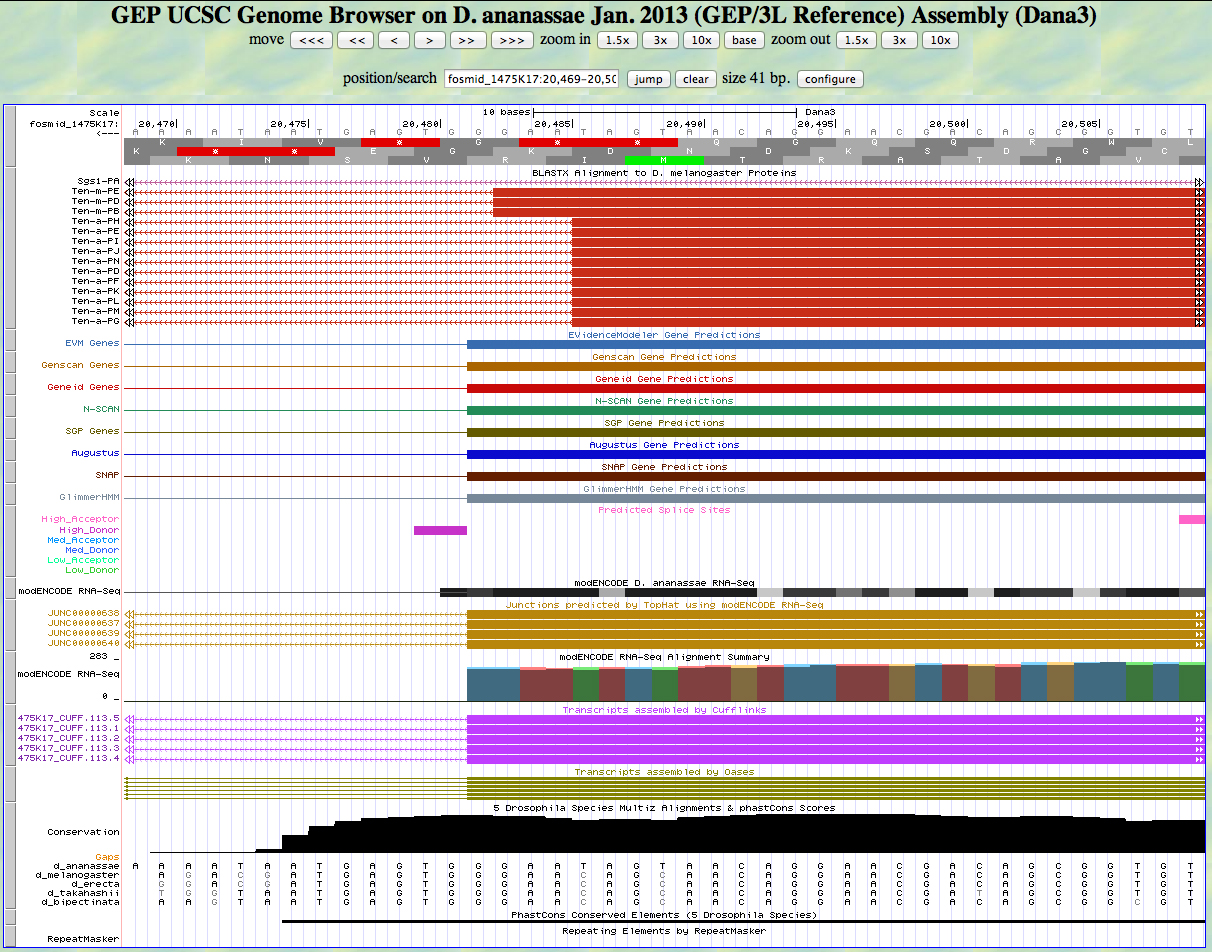
There is a GT predicted to be a high donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 20,482.
Here is the 5' end of the segment aligning to CDS 5 in the UCSC Genome Browser.
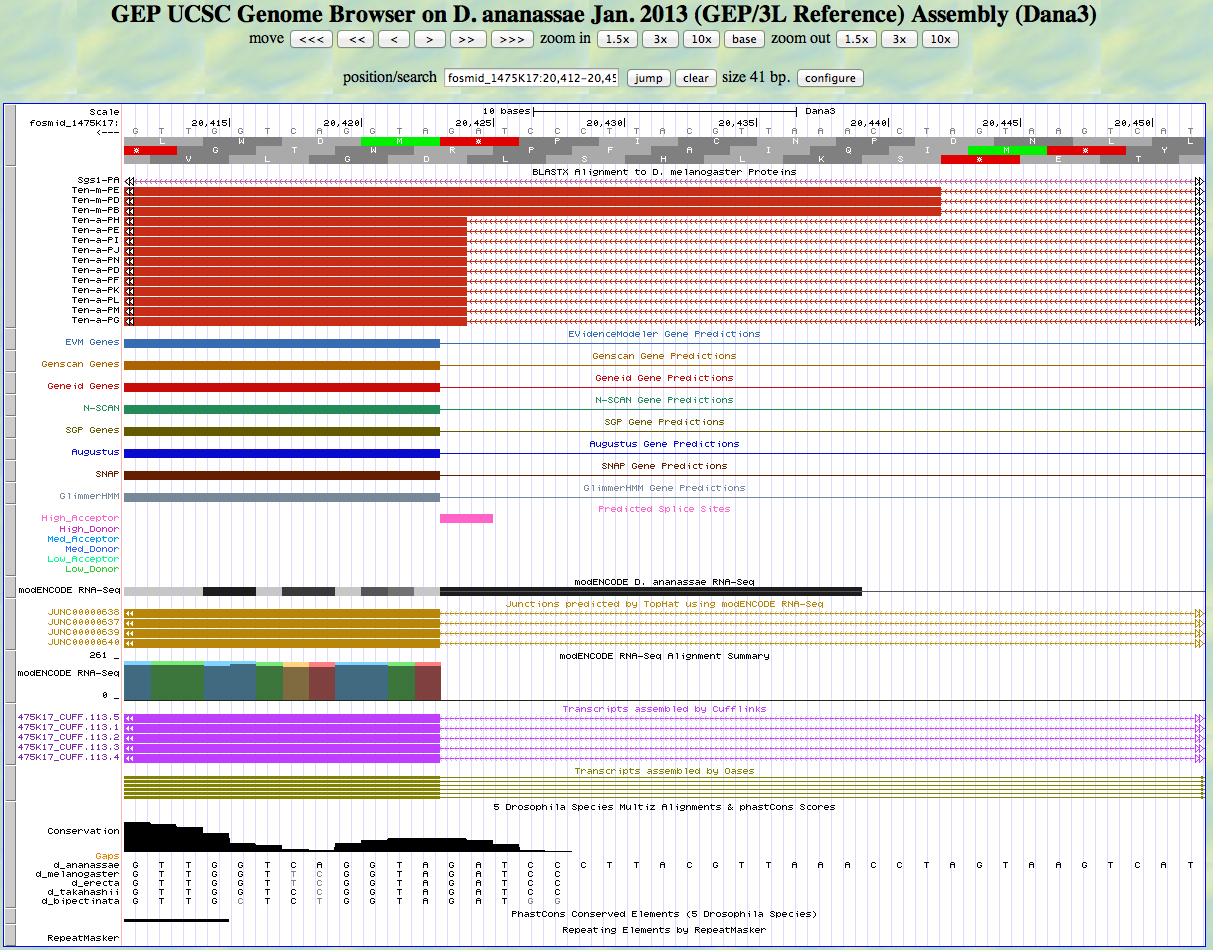
There is an AG predicted to be a high acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 20,423.
Here is the 3' end of the segment aligning to CDS 5 in the UCSC Genome Browser.
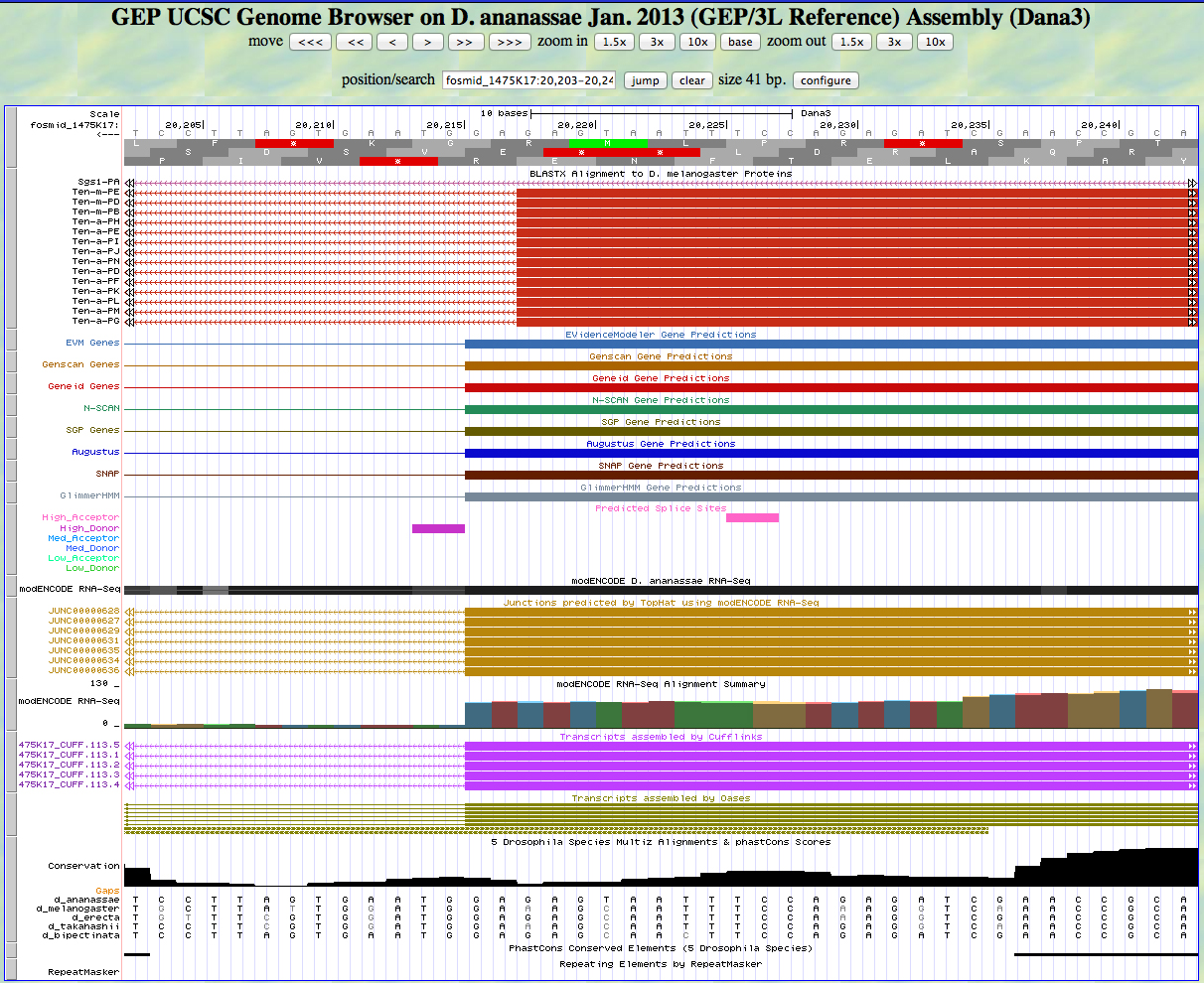
There is a GT predicted to be a high donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 20,216.
Here are the both ends of the segment aligning to CDS 4 in the UCSC Genome Browser.
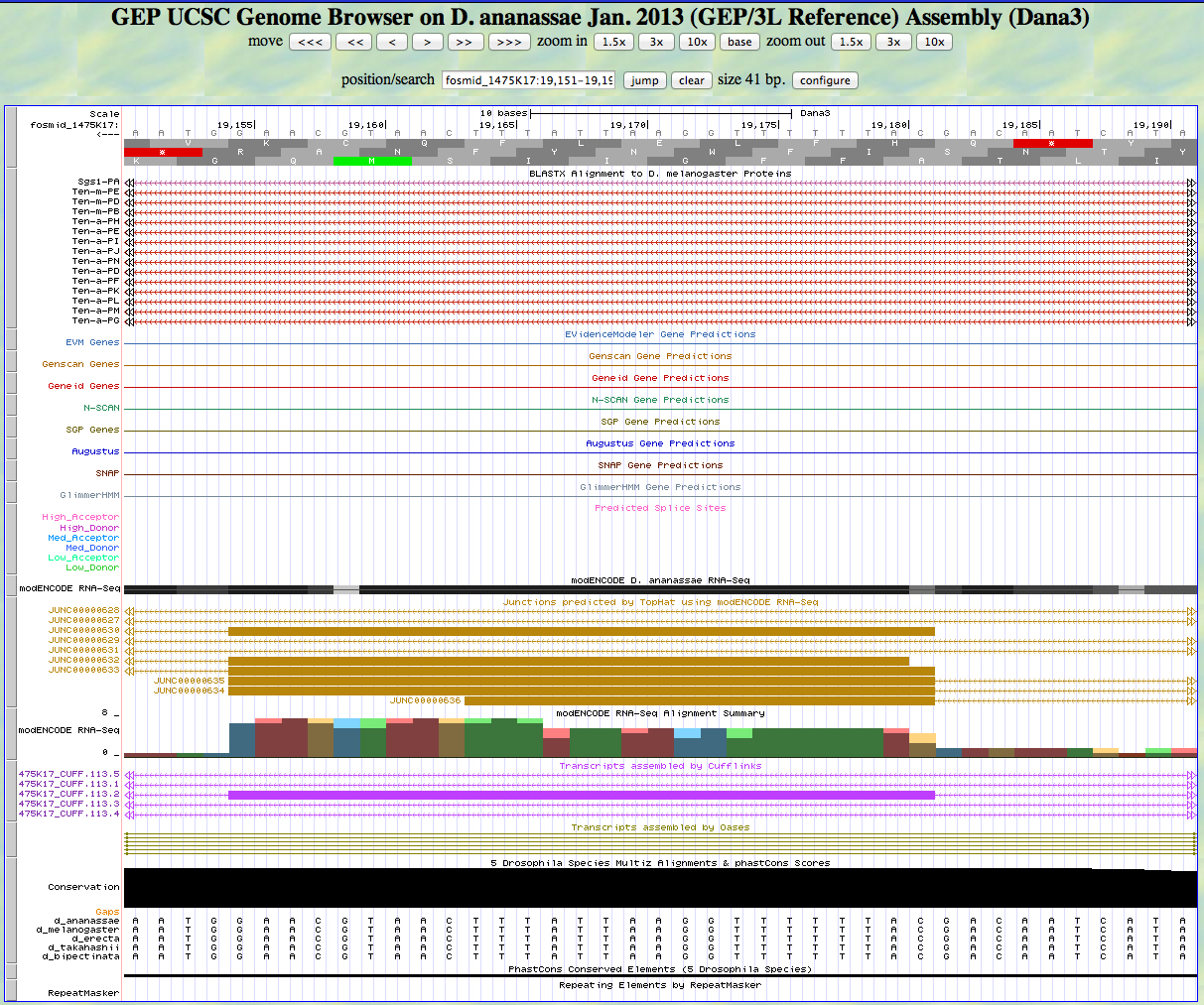
There is no alignment to D. melanogaster proteins displayed. Our BLASTX analysis gave an alignment to the eight amino acid CDS 4 at this approximate position with a score that was not significant. The gene finders all miss this segment also, but RNA-Seq (TopHat) data show this to be a real exon. The sequence conservation across five species is absolute.
There is an AG not predicted to be an acceptor at the position identified as a splice acceptor by RNA-Seq (TopHat). The first base of this exon is 19,181.
There is a GT not predicted to be a donor at the position identified as a splice donor by RNA-Seq (TopHat). The last base of this exon is 19,155.
Here is the 5' end of the segment aligning to CDS 3 in the UCSC Genome Browser.
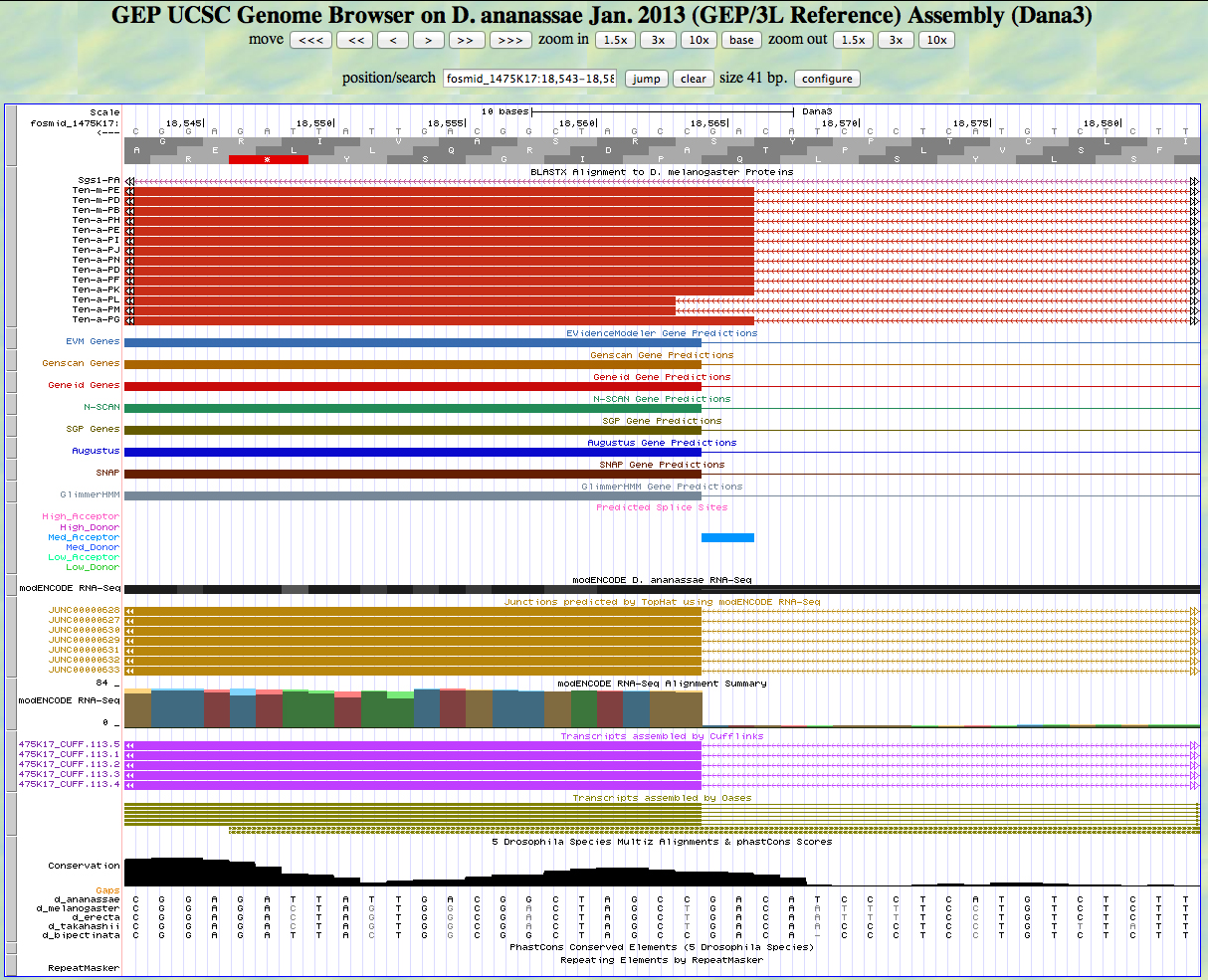
There is an AG predicted to be a medium acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 18,564.
Here is the 3' end of the segment aligning to CDS 3 in the UCSC Genome Browser.
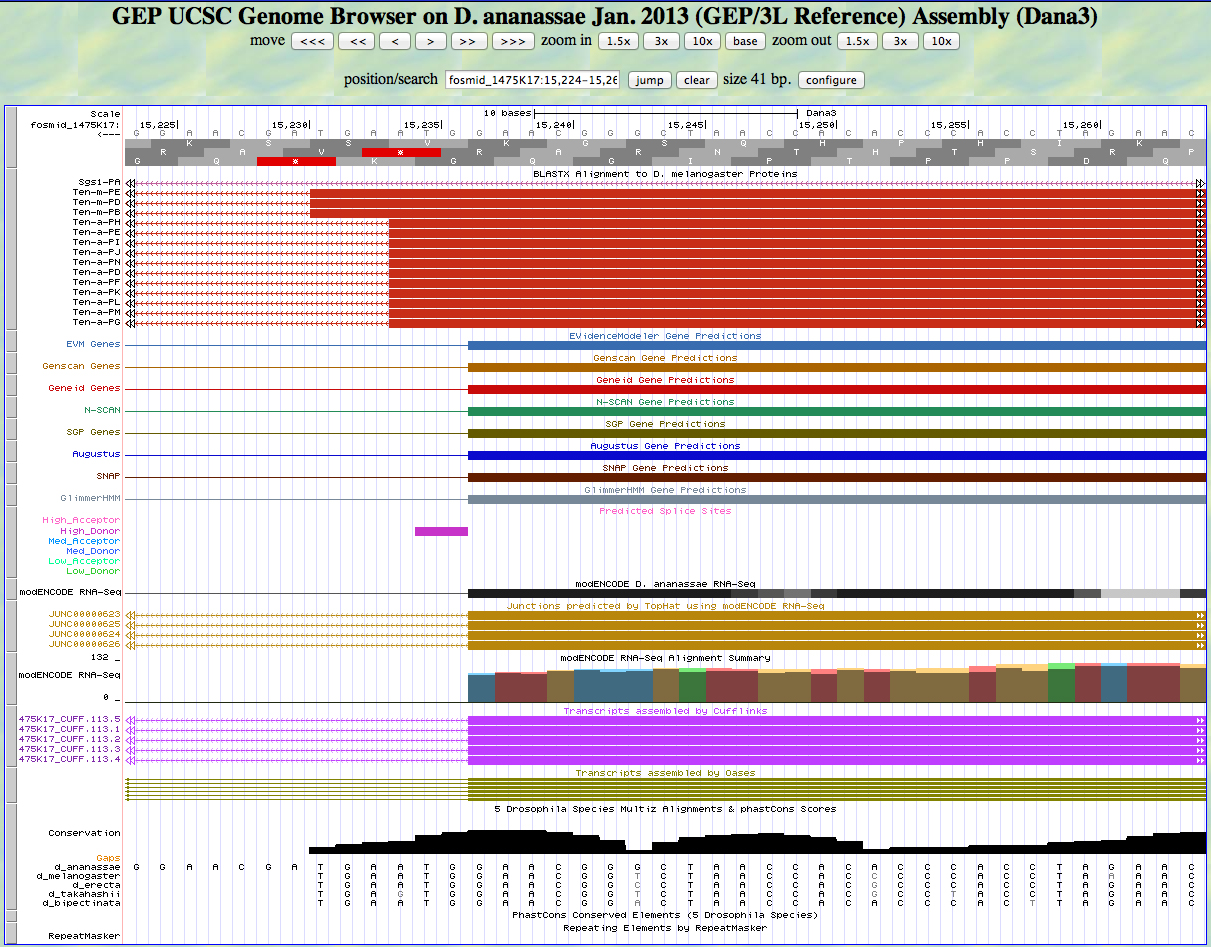
There is a GT predicted to be a high donor at the end of the alignment to the D. melanogaster protein, matching the 3' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The last base of this exon is 15,237.
Here is the 5' end of the segment aligning to CDS 1 (the same as for CDS 2) in the UCSC Genome Browser.
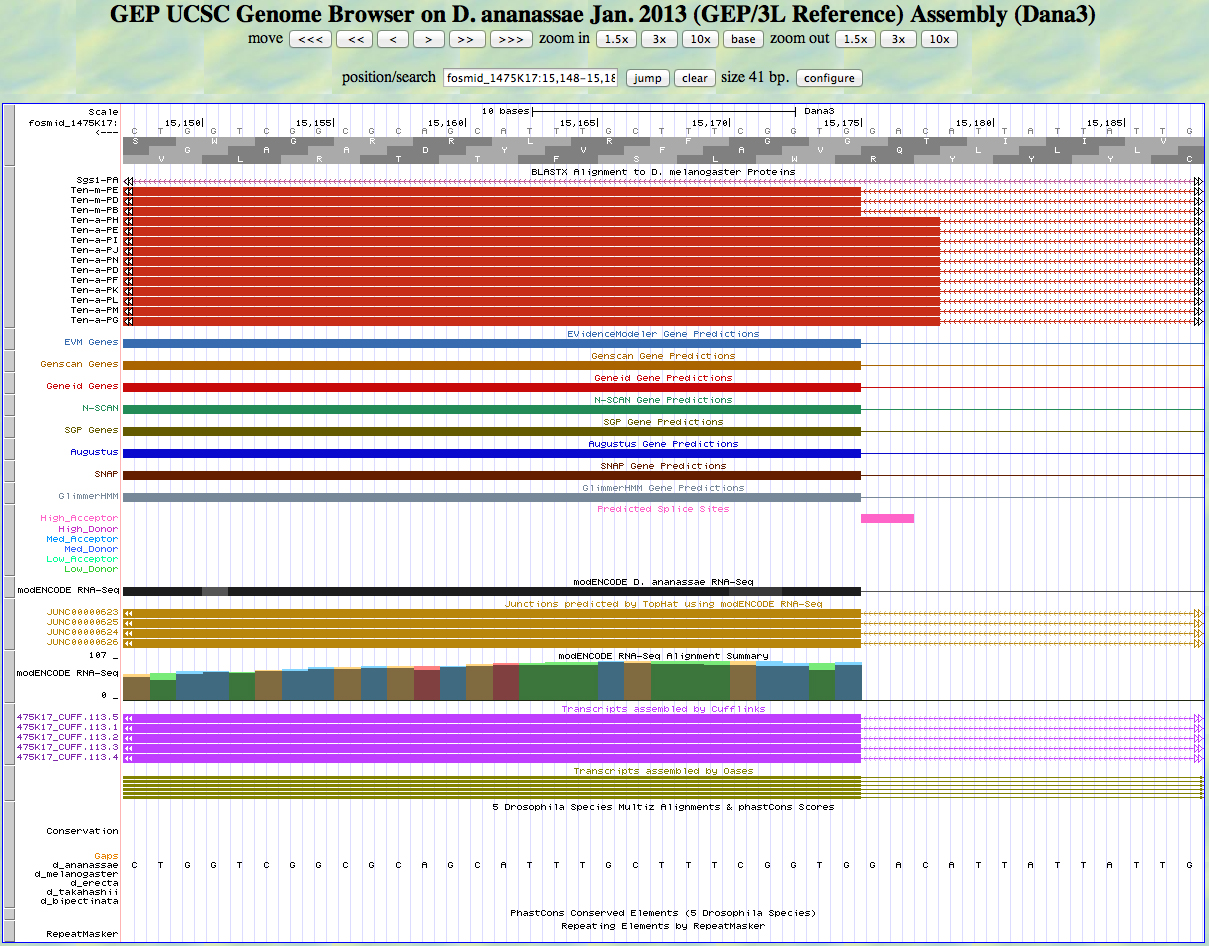
There is an AG predicted to be a high acceptor at the end of the alignment to the D. melanogaster protein, matching the 5' end predicted by all gene finders. The RNA-Seq data (TopHat) supports this position as well. The first base of this exon is 15,175.
Here is the 3' end of the segment aligning to CDS 1 in the UCSC Genome Browser.
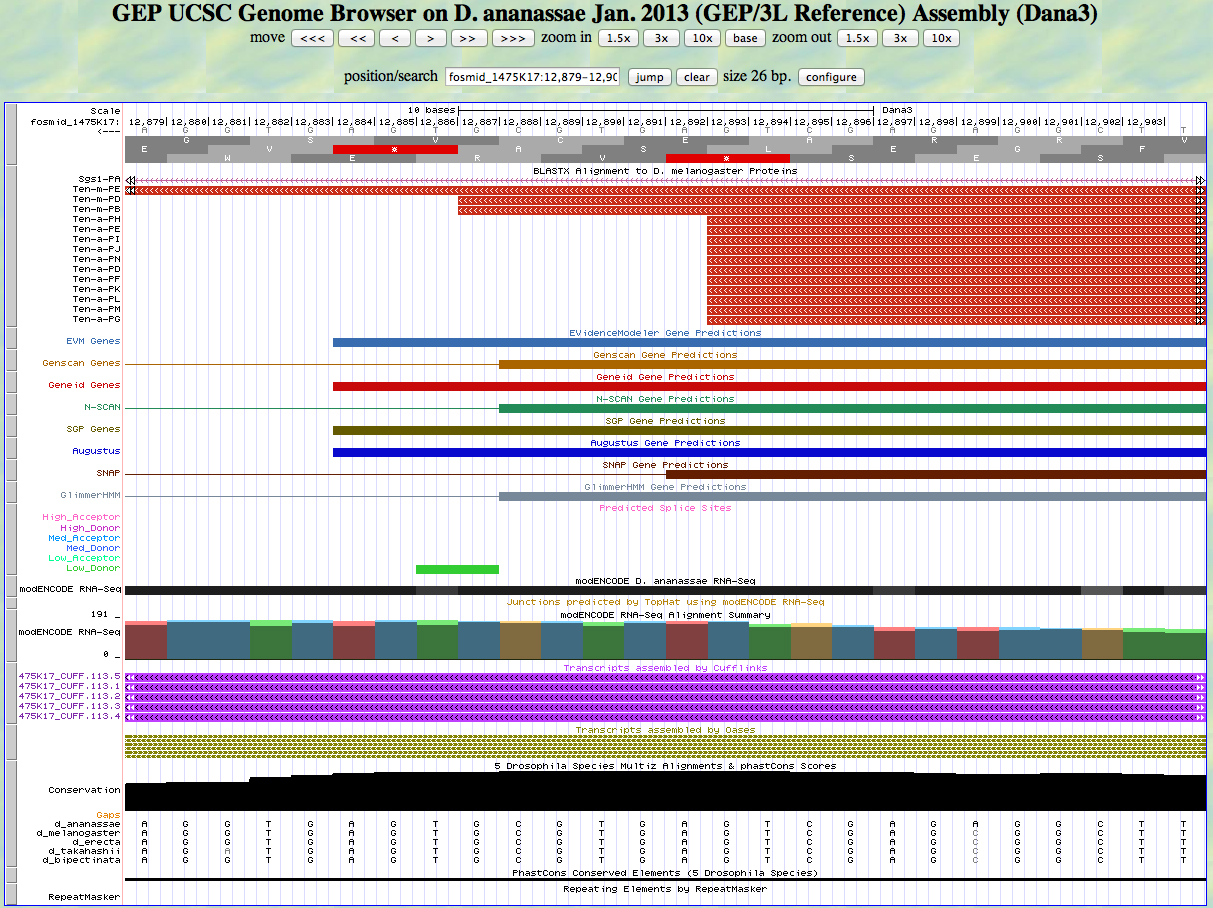
The last base of this exon is 12,887. The coordinates of the stop codon are 12,886 - 12,884.
Here is the 3' end of the segment aligning to CDS 2 in the UCSC Genome Browser.
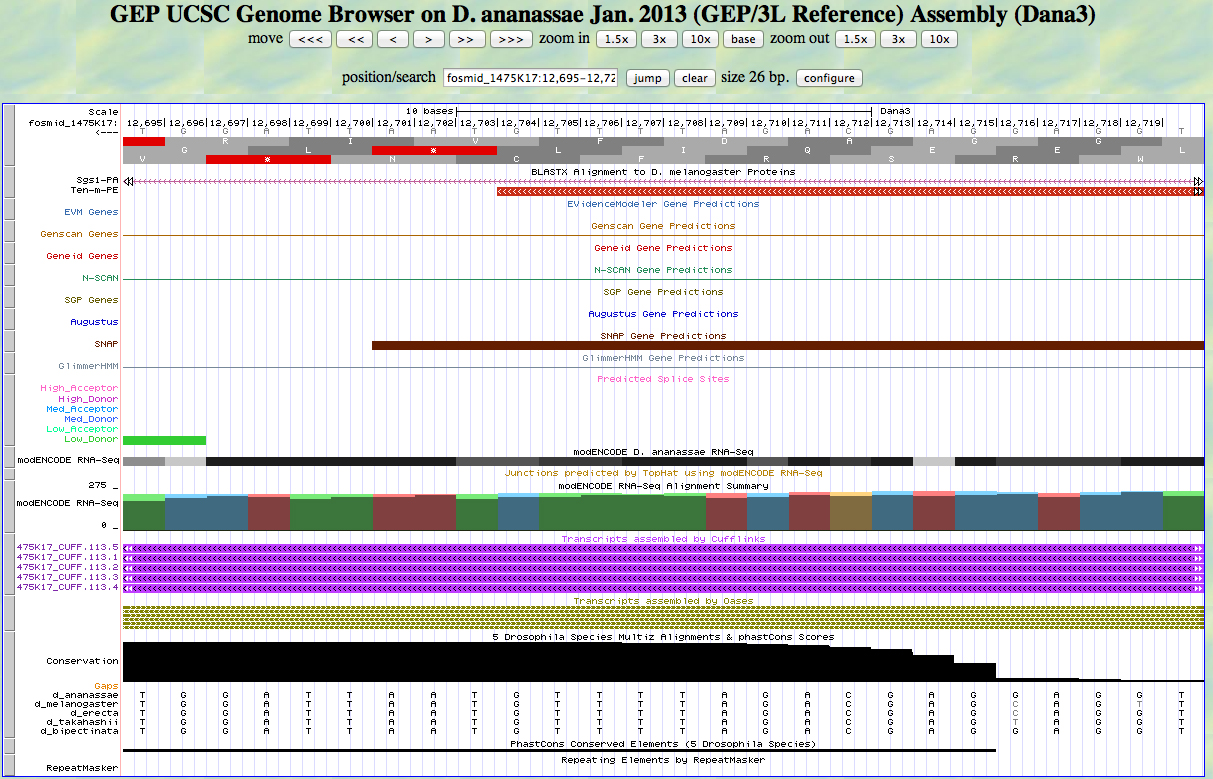
The last base of this exon is 12,704. The coordinates of the stop codon are 12,703 - 12,701.
| D. melanogaster Ten-m CDS segments | BLASTX | Model | ||||||||
| fosmid coords | alignment | |||||||||
| start | end | frame | E | identity | positive | phase | start | end | phase of next exon |
|
| Ten-m:11_536_2 | 25,756 | 25,316 | -2 | 7e-66 | 80% | 86% | 0 | 25,758 | 25,120 | 0 |
| Ten-m:10_602_2 | 24,496 | 23,612 | -2 | 2e-54 | 48% | 62% | 0 | 24,498 | 23,611 | 0 |
| Ten-m:9_602_2 | 23,192 | 22,365 | -1 | 6e-68 | 59% | 68% | 0 | 23,248 or 23,200 |
22,364 | 2 |
| Ten-m:8_536_2 | 21,938 | 21,657 | -1 | 1e-60 | 99% | 100% | 2 | 21,940 | 21,657 | 0 |
| Ten-m:7_536_0 | 21,528 | 21,118 | -3 | 2e-92 | 98% | 99% | 0 | 21,528 | 21,117 | 2 |
| Ten-m:6_536_2 | 21,025 | 20,483 | -2 | 2e-114 | 99% | 99% | 2 | 21,027 | 20,482 | 2 |
| Ten-m:5_536_2 | 20,421 | 20,218 | -3 | 1e-42 | 100% | 100% | 2 | 20,423 | 20,216 | 1 |
| Ten-m:4_602_1 | 19,180 | 19,157 | -2 | 0.036 | 100% | 100% | 1 | 19,181 | 19,155 | 1 |
| Ten-m:3_536_1 | 18,563 | 15,240 | -1 | 0.0 | 98% | 99% | 1 | 18,564 | 15,237 | 0 |
| Ten-m:1_536_0 | 15,175 | 12,884 | -2 | 0.0 | 96% | 98% | 0 | 15,175 | 12,887 | N/A |
| Ten-m:2_602_0 | 15,175 | 12,701 | -2 | 0.0 | 92% | 95% | 0 | 15,175 | 12,704 | N/A |
We will use the Gene Model Checker to determine if one of the two 5' ends for CDS 9 is better. First, here is the summary of CDS usage by isoform from the Gene Record Finder:
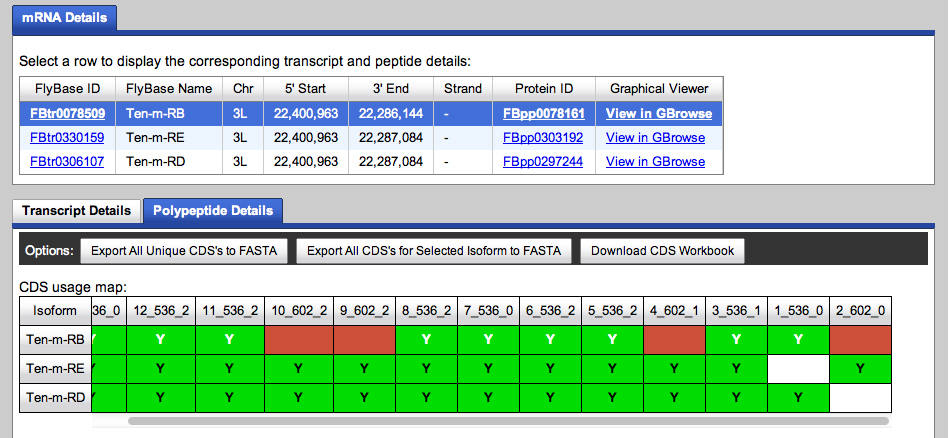
We will use Ten-m-PD to check the model. Ten-m-PB lacks CDS segments CDS 10 and CDS 9. Ten-m-PE has an unusual terminal coding segment (CDS 2) in which the stop codon that ends CDS 1 is read through.
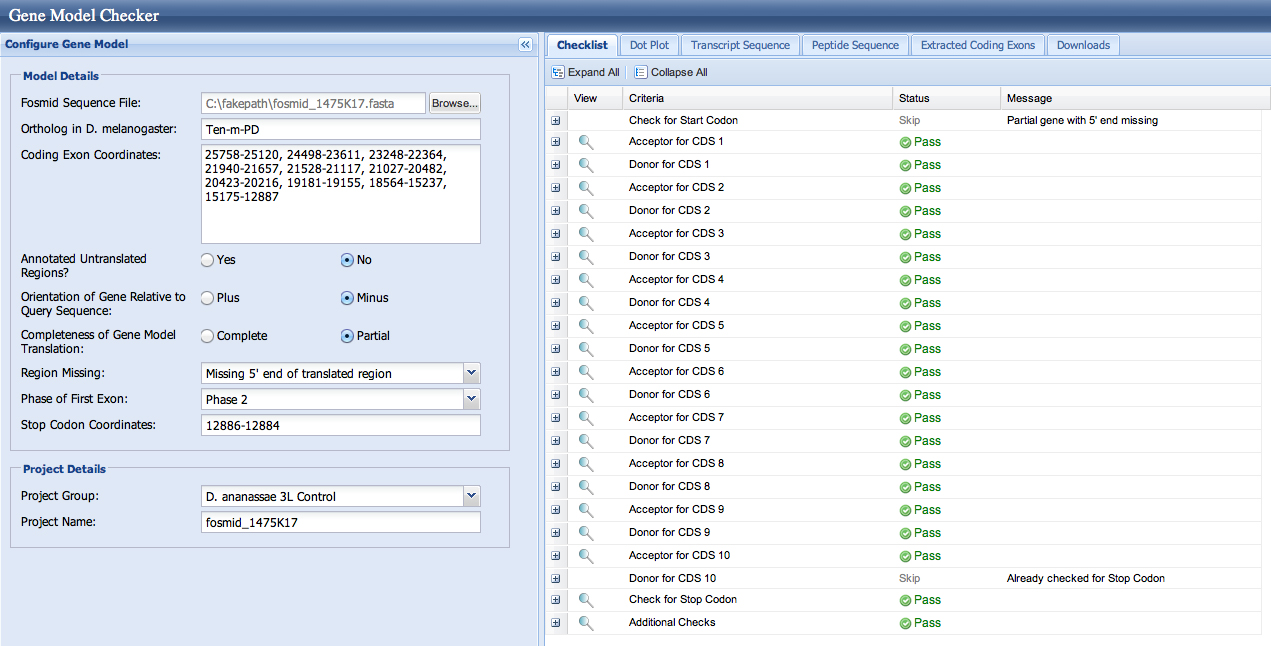
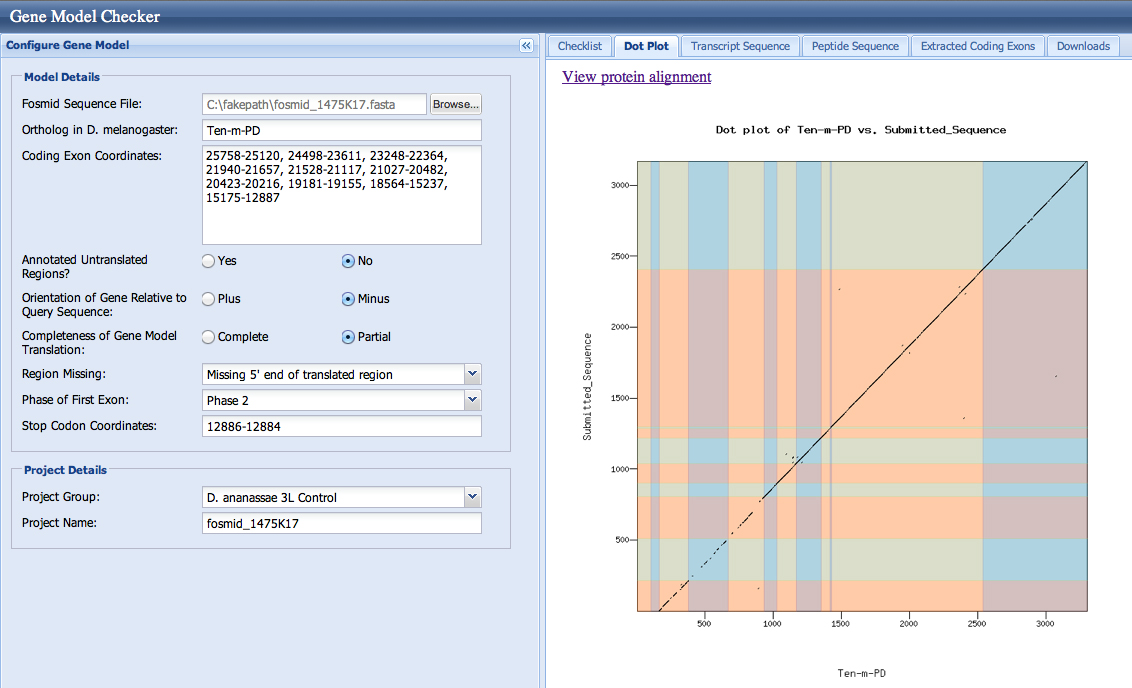
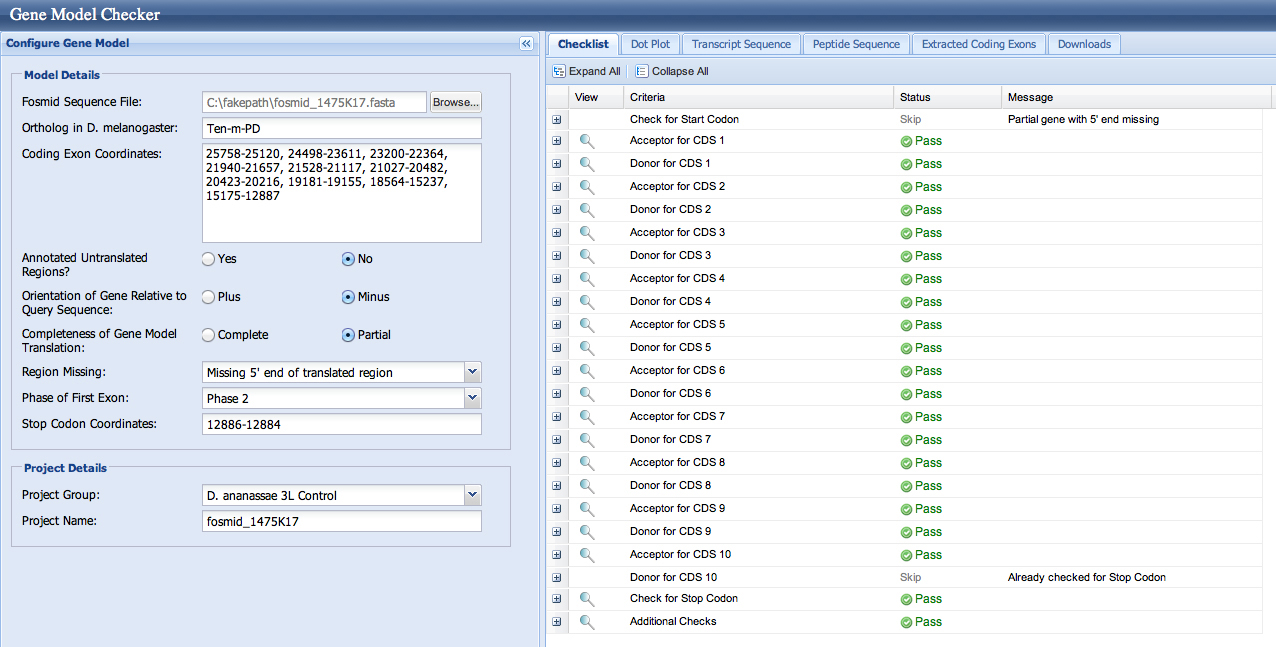
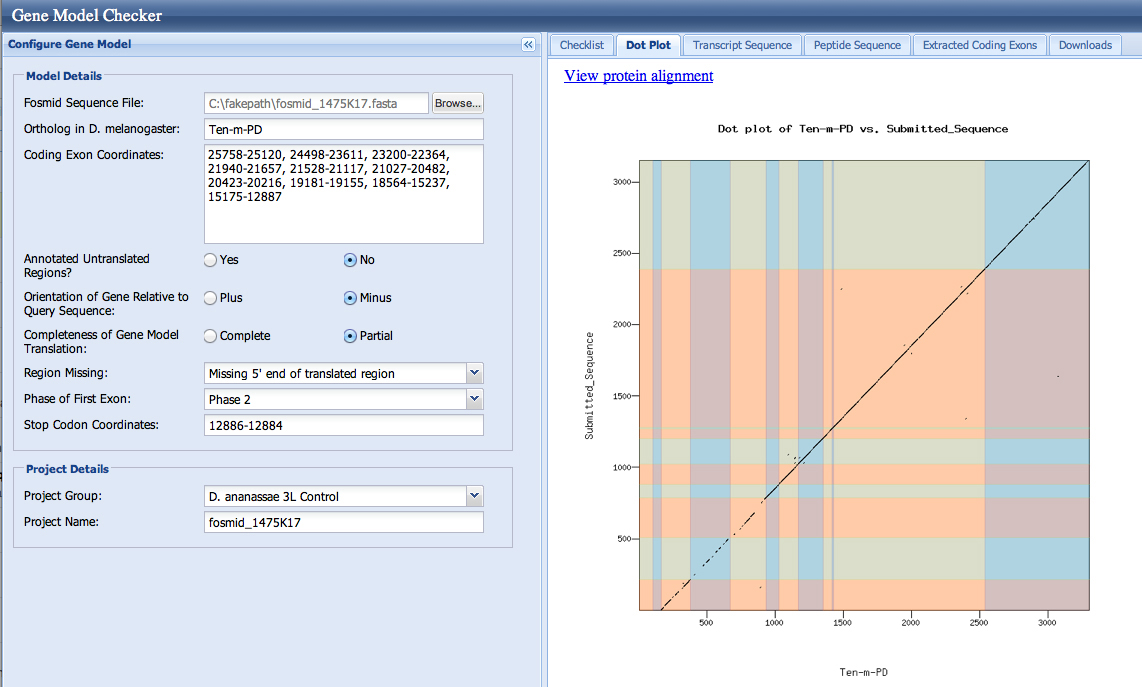
The checklist in the Gene Model Checker and the dotplots are good for both models. This is not surprising, as both splice acceptors (resulting in first coding base 23,248 and 23,200) are in phase 0.
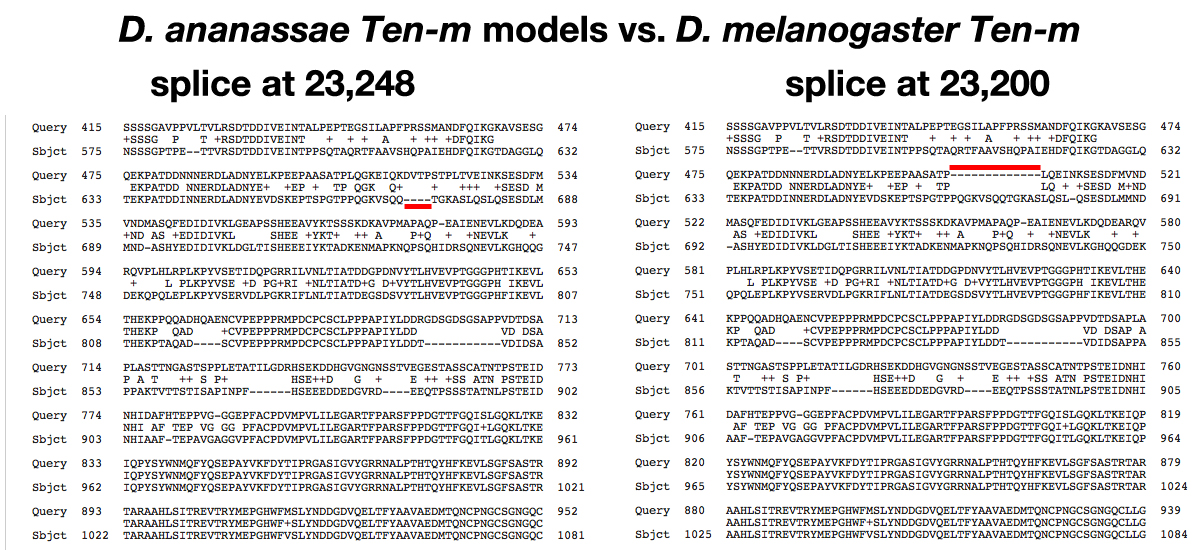
Both sequences aligned to NP_001262211.1. In the alignments, the longer form (splice at 23,248) requires a four amino acid gap to be introduced into the D. melanogaster protein to optimize the alignment. The shorter form (splice at 23,200) requires a thirteen amino acid gap to be introduced into the D. ananassae protein to optimize the alignment.
There is RNA-Seq evidence that both splice acceptors are used.
Given that isoform B (Ten-m-PB) entirely omits CDS 10 and CDS 9, these segments do not seem to be critical for protein function, so it is likely that both isoforms are made in D. ananassae. The complete protein sequences are given below.
>Ten-m-PD_peptide INKGSPIDFKSGSACSTPTKETLKGGYDRGTQGCMGPVLPPRSVMNGLPSHHYSAPMNFR KDLAARCSSPWVGVAAISALVLVVLMLILLTTFGGLHWTQSAPCTVLVGNEASEVTAAKS TNTDLSKLHNSATRSKNGQAIGPVPGQYGSGGGMGSSTATVTAATSNSGTAQGLQSTSAS AEASSSAATTSSQSSLTPSTSLSSSLANANNGGRDILTRMAADGAGKNKRRNRRSMDVAE NGGDVATDETFSNFITIESLNREQTGEYFATTPARKLQEVERSSSDRTSFGINGVLSPQG DEEVEDITSDYVYEDEPVPDTSPATQRPRTRQQFGKSLNSNLRSAAKTLVNKKTKYEGEA GKNIRLEQEQKLEAFIEAGMTLESTTTTATRATTTTESGTSTFVAVIDDDNQSDSSSSGA VPPVLTVLRSDTDDIVEINTALPEPTEGSILAPFPRSSMANDFQIKGKAVSESGQEKPAT DDNNNERDLADNYELKPEEPAASATPLQGKEIQKDVTPSTPLTVEINKSESDFMVNDMAS QFEDIDIVKLGEAPSSHEEAVYKTSSSKDKAVPMAPAQPEAIENEVLKDQDEARQVPLHL RPLKPYVSETIDQPGRRILVNLTIATDDGPDNVYTLHVEVPTGGGPHTIKEVLTHEKPPQ QADHQAENCVPEPPPRMPDCPCSCLPPPAPIYLDDRGDSGDSGSAPPVDTDSAPLASTTN GASTSPPLETATILGDRHSEKDDHGVGNGNSSTVEGESTASSCATNTPSTEIDNHIDAFH TEPPVGGGEPFACPDVMPVLILEGARTFPARSFPPDGTTFGQISLGQKLTKEIQPYSYWN MQFYQSEPAYVKFDYTIPRGASIGVYGRRNALPTHTQYHFKEVLSGFSASTRTARAAHLS ITREVTRYMEPGHWFMSLYNDDGDVQELTFYAAVAEDMTQNCPNGCSGNGQCLLGHCQCN PGFGGHDCSESVCPVLCSQHGEYTNGECICNPGWKGKECSLRHDECEVADCNGHGHCVSG KCQCMRGYKGKFCEEVDCPHPNCSGHGFCADGTCICKKGWKGPDCATMDQDALQCLPDCS GHGTFDLDTQTCTCEAKWSGDDCSKELCDLDCGQHGRCEGDACACDPEWGGEYCNTRLCD TRCNEHGQCKNGTCLCVTGWNGKHCTIEGCPNSCAGHGQCRVSGEGQWECRCYEGWDGPD CGIALELNCGDSKDNDKDGLVDCEDPECCASHVCKTSQLCVSAPKPIDVLLRKQPPAITA SFFERMKFLIDESSLQNYAKLETFNESIFWNYFNASRSAVIRGRVVTSLGMGLVGVRVST TTLLEGFTLTRDDGWFDLMVNGGGAVTLQFGRAPFRPQSRIVQVPWNEVVIIDGVVMSMS EEKGLATTTTHTCFAHDYDLMKPVVLASWKHGFQGACPDRSAILAESQVIQESLQIPGTG LNLVYHSSRAAGYLSTIKLQLTPDNIPPTLHLIHLRITIEGILFERVFEADPGIKFTYAW NRLNIYRQRVYGVTTAVVKVGYQYTDCTDIVWDIQTTKLSGHDMSISEVGGWNLDIHHRY NFHEGILQKGDGSNIYLRNKPRIILTTMGDGHQRPLECPDCDGLATKQRLLAPVALAAAP DGSLFVGDFNYIRRIMSDGSIRTVVKLNATRVSYRYHMALSPLDGTLYVSDPESHQIIRV RDTNNYSQPELNWEAVVGSGERCLPGDEAHCGDGALAKDAKLAYPKGIAISSDNILYFAD GTNIRMVDRDGIVSTLIGNHMHKSHWKPIPCEGTLKLEEMHLRWPTELAVSPMDNTLHII DDHMILRMTPDGRVRVISGRPLHCATASTAYDTDLATHATLVMPQSIAFGPLGELYVAES DSQRINRVRVIGTDGRIAPFAGAESKCNCLERGCDCFEAEHYLATSAKFNTIAALSVTPD GHVHIADQANYRIRSVMSSIPEASPSREYEIYAPDMQEIYIFNRFGQHVSTRNILTGETT YVFTYNVNTSNGKLSTVTDAAGNKVFLLRDYTSQVNSIENTKGQKCRLRMTRMKMLHELS TPDNYNVTYEYHGPTGLLKTKLDSTGRSYVYNYDEFGRLTSAVTPTGRVIELSFDLSVKG AQVKVSENAQKEQSLLIQGATVTVRNGAAESRTSVDMDGSTTSITPWGHNVQMEVAPYTI LAEQSPLLGESYPVPAKQRTEIAGDLANRFEWRYFVRRQQPLQAGKQSKGAPRPVTEVGR KLRVNGDNVLTLEYDRETQSVVVLVDDKQELLNVTYDRTSRPISFRPQSGDYADVDLEYD RFGRLVSWKWGVLQEAYSFDRNGRLNEIKYGDGSTMVYAFKDMFGSLPLKVTTPRRSDYL LQYDDAGALQSLTTPRGHIHAFSLQTSLGFFKYQYFSPINRHPFEILYNDEGQILAKIHP HQSGKVAFVYDAAGRLETILAGLSSTHYTYQDTTSLVKTVEVQEPGFELRREFKYHAGIL KDEKLRFGSKNSLASAHYKYAYDGNARLSGIEMAIDDKELPTTRYKYSQNLGQLEVVQDL KITRNAFNRTVIQDSAKQFFAIVDYDQHGRVKSVLMNVKNIDVFRLELDYDLRNRIKSQK TTFGRSTAFDKINYNADGHVVEVLGTNNWKYLYDENGNTVGVVDQGEKFNLGYDIGDRVI KVGDVEFNNYDARGFVVRRGEQKYRYNNRGQLIHAFERERFQSWYYYDDRSRLVAWHDNQ GNTTQYYYANPRTPHLVTHAHFPKLARTMKFFYDDRDMLIAMENADQRYYVATDQNGSPL AFFDLNGGIAKELKRTPFGRIIKDTKPDFFVPIDFHGGLIDPHTKLIYTEQRQYDPHVGQ WMTPQWETLATEMSHPTDVFIYRYHNNDPINPNRPQNYMIDLDAWLQLFGYDLDNMQSRR YTKLAQYTPQASIKSNMLAPDFGVISGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLL PRISYRRGVFGEGVLLSRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDV FYFVKDNVLKLRDDNEELRRLGGMFNISTHEVSDHGGSAAKELRLHGPDAVVIVKYGVDP EQERHRILKHAHKRAVERAWELEKQLVAAGFQGRGDWTEEEKEELVQHGDVDGWIGIDIH SIHKYPQLADDPGNVAFQRDAKRKRRKTGNSHRSASSRRQMKFGELSA >Ten-m-PD_peptide INKGSPIDFKSGSACSTPTKETLKGGYDRGTQGCMGPVLPPRSVMNGLPSHHYSAPMNFR KDLAARCSSPWVGVAAISALVLVVLMLILLTTFGGLHWTQSAPCTVLVGNEASEVTAAKS TNTDLSKLHNSATRSKNGQAIGPVPGQYGSGGGMGSSTATVTAATSNSGTAQGLQSTSAS AEASSSAATTSSQSSLTPSTSLSSSLANANNGGRDILTRMAADGAGKNKRRNRRSMDVAE NGGDVATDETFSNFITIESLNREQTGEYFATTPARKLQEVERSSSDRTSFGINGVLSPQG DEEVEDITSDYVYEDEPVPDTSPATQRPRTRQQFGKSLNSNLRSAAKTLVNKKTKYEGEA GKNIRLEQEQKLEAFIEAGMTLESTTTTATRATTTTESGTSTFVAVIDDDNQSDSSSSGA VPPVLTVLRSDTDDIVEINTALPEPTEGSILAPFPRSSMANDFQIKGKAVSESGQEKPAT DDNNNERDLADNYELKPEEPAASATPLQEINKSESDFMVNDMASQFEDIDIVKLGEAPSS HEEAVYKTSSSKDKAVPMAPAQPEAIENEVLKDQDEARQVPLHLRPLKPYVSETIDQPGR RILVNLTIATDDGPDNVYTLHVEVPTGGGPHTIKEVLTHEKPPQQADHQAENCVPEPPPR MPDCPCSCLPPPAPIYLDDRGDSGDSGSAPPVDTDSAPLASTTNGASTSPPLETATILGD RHSEKDDHGVGNGNSSTVEGESTASSCATNTPSTEIDNHIDAFHTEPPVGGGEPFACPDV MPVLILEGARTFPARSFPPDGTTFGQISLGQKLTKEIQPYSYWNMQFYQSEPAYVKFDYT IPRGASIGVYGRRNALPTHTQYHFKEVLSGFSASTRTARAAHLSITREVTRYMEPGHWFM SLYNDDGDVQELTFYAAVAEDMTQNCPNGCSGNGQCLLGHCQCNPGFGGHDCSESVCPVL CSQHGEYTNGECICNPGWKGKECSLRHDECEVADCNGHGHCVSGKCQCMRGYKGKFCEEV DCPHPNCSGHGFCADGTCICKKGWKGPDCATMDQDALQCLPDCSGHGTFDLDTQTCTCEA KWSGDDCSKELCDLDCGQHGRCEGDACACDPEWGGEYCNTRLCDTRCNEHGQCKNGTCLC VTGWNGKHCTIEGCPNSCAGHGQCRVSGEGQWECRCYEGWDGPDCGIALELNCGDSKDND KDGLVDCEDPECCASHVCKTSQLCVSAPKPIDVLLRKQPPAITASFFERMKFLIDESSLQ NYAKLETFNESIFWNYFNASRSAVIRGRVVTSLGMGLVGVRVSTTTLLEGFTLTRDDGWF DLMVNGGGAVTLQFGRAPFRPQSRIVQVPWNEVVIIDGVVMSMSEEKGLATTTTHTCFAH DYDLMKPVVLASWKHGFQGACPDRSAILAESQVIQESLQIPGTGLNLVYHSSRAAGYLST IKLQLTPDNIPPTLHLIHLRITIEGILFERVFEADPGIKFTYAWNRLNIYRQRVYGVTTA VVKVGYQYTDCTDIVWDIQTTKLSGHDMSISEVGGWNLDIHHRYNFHEGILQKGDGSNIY LRNKPRIILTTMGDGHQRPLECPDCDGLATKQRLLAPVALAAAPDGSLFVGDFNYIRRIM SDGSIRTVVKLNATRVSYRYHMALSPLDGTLYVSDPESHQIIRVRDTNNYSQPELNWEAV VGSGERCLPGDEAHCGDGALAKDAKLAYPKGIAISSDNILYFADGTNIRMVDRDGIVSTL IGNHMHKSHWKPIPCEGTLKLEEMHLRWPTELAVSPMDNTLHIIDDHMILRMTPDGRVRV ISGRPLHCATASTAYDTDLATHATLVMPQSIAFGPLGELYVAESDSQRINRVRVIGTDGR IAPFAGAESKCNCLERGCDCFEAEHYLATSAKFNTIAALSVTPDGHVHIADQANYRIRSV MSSIPEASPSREYEIYAPDMQEIYIFNRFGQHVSTRNILTGETTYVFTYNVNTSNGKLST VTDAAGNKVFLLRDYTSQVNSIENTKGQKCRLRMTRMKMLHELSTPDNYNVTYEYHGPTG LLKTKLDSTGRSYVYNYDEFGRLTSAVTPTGRVIELSFDLSVKGAQVKVSENAQKEQSLL IQGATVTVRNGAAESRTSVDMDGSTTSITPWGHNVQMEVAPYTILAEQSPLLGESYPVPA KQRTEIAGDLANRFEWRYFVRRQQPLQAGKQSKGAPRPVTEVGRKLRVNGDNVLTLEYDR ETQSVVVLVDDKQELLNVTYDRTSRPISFRPQSGDYADVDLEYDRFGRLVSWKWGVLQEA YSFDRNGRLNEIKYGDGSTMVYAFKDMFGSLPLKVTTPRRSDYLLQYDDAGALQSLTTPR GHIHAFSLQTSLGFFKYQYFSPINRHPFEILYNDEGQILAKIHPHQSGKVAFVYDAAGRL ETILAGLSSTHYTYQDTTSLVKTVEVQEPGFELRREFKYHAGILKDEKLRFGSKNSLASA HYKYAYDGNARLSGIEMAIDDKELPTTRYKYSQNLGQLEVVQDLKITRNAFNRTVIQDSA KQFFAIVDYDQHGRVKSVLMNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNA DGHVVEVLGTNNWKYLYDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGFV VRRGEQKYRYNNRGQLIHAFERERFQSWYYYDDRSRLVAWHDNQGNTTQYYYANPRTPHL VTHAHFPKLARTMKFFYDDRDMLIAMENADQRYYVATDQNGSPLAFFDLNGGIAKELKRT PFGRIIKDTKPDFFVPIDFHGGLIDPHTKLIYTEQRQYDPHVGQWMTPQWETLATEMSHP TDVFIYRYHNNDPINPNRPQNYMIDLDAWLQLFGYDLDNMQSRRYTKLAQYTPQASIKSN MLAPDFGVISGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRISYRRGVFGEGVLL SRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVKDNVLKLRDDNE ELRRLGGMFNISTHEVSDHGGSAAKELRLHGPDAVVIVKYGVDPEQERHRILKHAHKRAV ERAWELEKQLVAAGFQGRGDWTEEEKEELVQHGDVDGWIGIDIHSIHKYPQLADDPGNVA FQRDAKRKRRKTGNSHRSASSRRQMKFGELSA
We would like to know if the two different splice acceptors predicted at the beginning of D. ananassae CDS 9 are possible in D. melanogaster. Decorated genomic sequences are presented below.
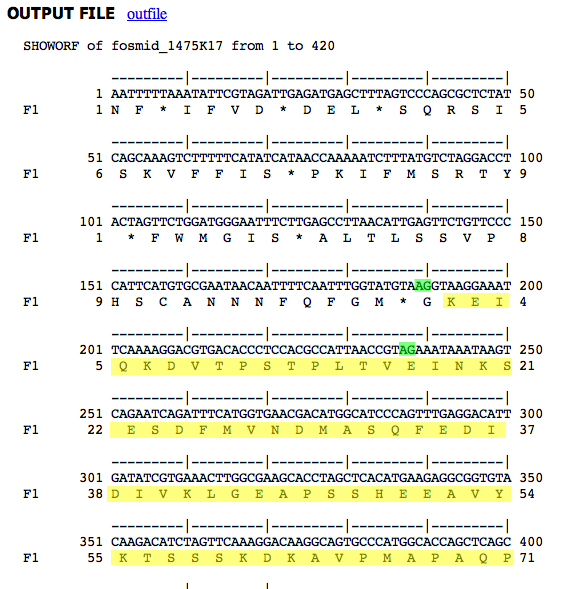 |
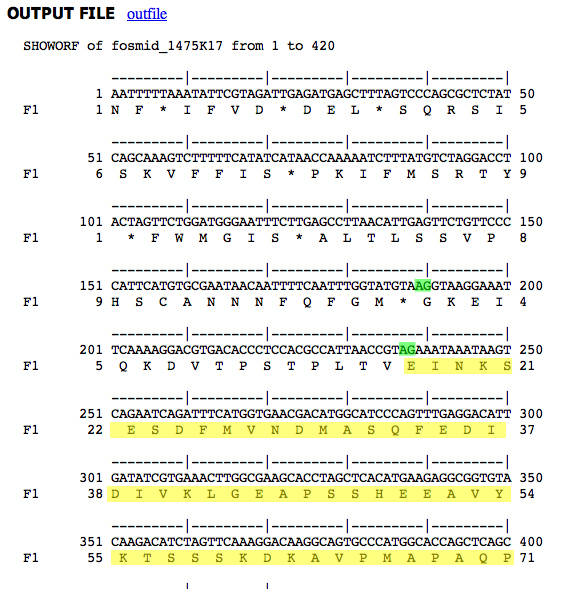 |
 |
| D. ananassae genomic sequence Splice acceptors in green, CDS 9 peptide in yellow. First splice acceptor used. |
D. ananassae genomic sequence Splice acceptors in green, CDS 9 peptide in yellow. Second splice acceptor used. |
D. melanogaster genomic sequence Splice acceptor in green, CDS 9 peptide in yellow. First splice acceptor used, no second acceptor. |
It appears that the splice acceptor that makes the shorter isoform possible in D. ananassae is not present in D. melanogaster.