| Summary | |
| BLASTX Analysis Graphic summary | |
| BLASTX Analysis Descriptions | |
| BLASTX Analysis Ten-m alignment | |
| BLASTX Analysis Hem alignment | |
| BLASTX Analysis Aats-ile alignment | |
| BLAST Analysis by GEP | |
| BLASTX Analysis at FlyBase | |
| GENSCAN Analysis | |
| UCSC Genome Browser at GEP | |
| Lightning Round Slides |
D. ananassae fosmid 1475K17 encodes three genes: the D. ananassae orthologs of the D. melanogaster genes Hem, Aats-ile, and Ten-m. The Hem and Aats-ile genes are complete, while the 5' end of the Ten-m gene is not on the fosmid. Based on the gene model of the D. melanogaster Ten-m gene, the first and second exons are missing. GENSCAN predicts five peptides encoded on the fosmid. Three of these peptides match the three genes identified by BLASTX analysis, while two (peptides 4 and 5) are invalid predictions.
I used the file fosmid_1475K17.fasta from the src folder in the GEP project file for D. ananassae fosmid 1475K17 as a query sequence in a BLASTX search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings.
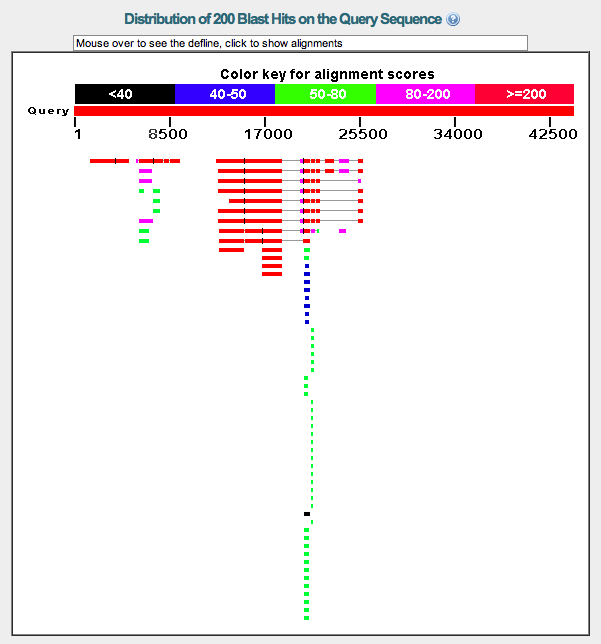
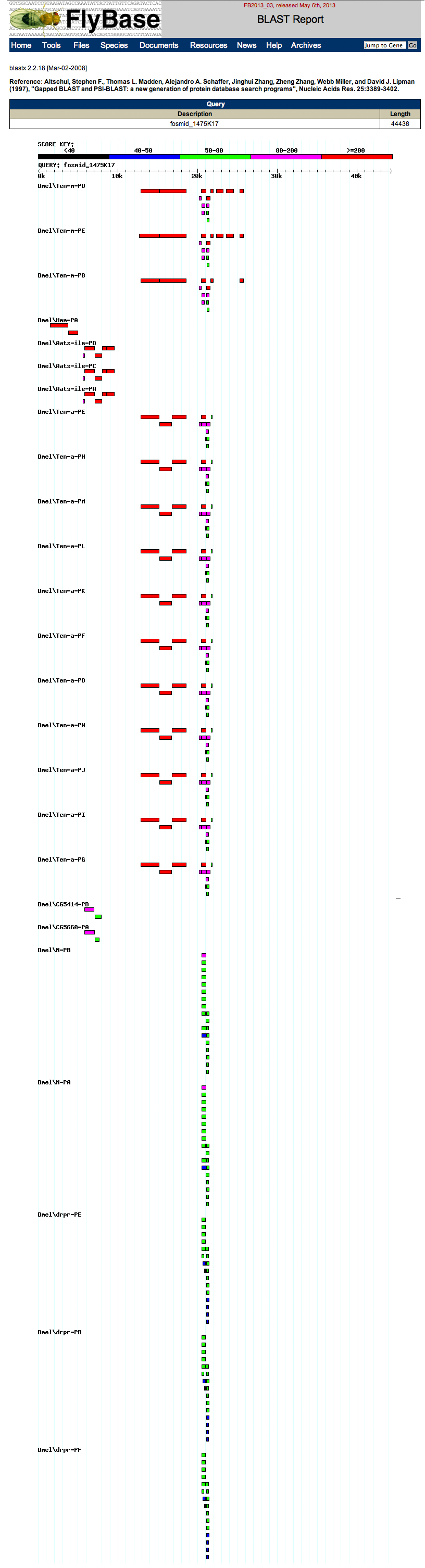
Graphic summary. The graphic summary is shown below.

The results suggest that there are three genes on the fosmid with protein sequence similarity to the D. melanogaster protein set.
The left gene (Gene A) has only one good match to D. melanogaster proteins.
The middle gene (Gene B) has one good match to D. melanogaster proteins, with eight additional hits at lower scores. Each of the eight lower-scoring hits covers only a portion of the top hit.
The right gene (Gene C) has multiple good matches. These appear to be isoforms derived from alternative splicing. There are many lower-scoring hits that cover only a portion of the top hit.
Descriptions. All descriptions with E values smaller than e-10 are shown below.
Alignments
Gene C. The first seven alignments in the list of descriptions (tenascin, odd Oz, odz) all align to a region of the fosmid corresponding to Gene C in the graphic summary. The coordinates of the top hit (tenascin major, isoform E) are shown in the table below.
| NP_001262211.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 1425 | 2533 | 18566 | 15240 | -1 | 0.0 | 98% | 99% |
| 2534 | 3349 | 15178 | 12704 | -2 | 0.0 | 92% | 95% |
| 1165 | 1346 | 21028 | 20483 | -2 | 0.0 | 99% | 99% |
| 1026 | 1164 | 21534 | 21118 | -3 | 0.0 | 97% | 98% |
| 933 | 1033 | 21941 | 21639 | -1 | 0.0 | 95% | 98% |
| 1341 | 1415 | 20442 | 20218 | -3 | 0.0 | 95% | 96% |
| 682 | 933 | 23192 | 22362 | -1 | 1e-57 | 59% | 68% |
| 161 | 307 | 25759 | 25316 | -2 | 6e-54 | 79% | 86% |
| 376 | 667 | 24499 | 23609 | -2 | 2e-46 | 49% | 62% |
| 1086 | 1195 | 20860 | 20528 | -2 | 6e-11 | 39% | 52% |
| 1237 | 1330 | 21399 | 21124 | -3 | 1e-09 | 39% | 54% |
| 1140 | 1234 | 21387 | 21118 | -3 | 2e-08 | 40% | 53% |
| 1053 | 1162 | 20872 | 20531 | -2 | 6e-06 | 35% | 50% |
| 1265 | 1332 | 21408 | 21220 | -3 | 0.054 | 35% | 48% |
Reordering these in order of the segments of NP_001262211.1 gives the results shown below. Alignments with E values larger than e-10 are grayed out.
| NP_001262211.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 161 | 307 | 25759 | 25316 | -2 | 6e-54 | 79% | 86% |
| 376 | 667 | 24499 | 23609 | -2 | 2e-46 | 49% | 62% |
| 682 | 933 | 23192 | 22362 | -1 | 1e-57 | 59% | 68% |
| 933 | 1033 | 21941 | 21639 | -1 | 0.0 | 95% | 98% |
| 1026 | 1164 | 21534 | 21118 | -3 | 0.0 | 97% | 98% |
| 1053 | 1162 | 20872 | 20531 | -2 | 6e-06 | 35% | 50% |
| 1086 | 1195 | 20860 | 20528 | -2 | 6e-11 | 39% | 52% |
| 1140 | 1234 | 21387 | 21118 | -3 | 2e-08 | 40% | 53% |
| 1165 | 1346 | 21028 | 20483 | -2 | 0.0 | 99% | 99% |
| 1237 | 1330 | 21399 | 21124 | -3 | 1e-09 | 39% | 54% |
| 1265 | 1332 | 21408 | 21220 | -3 | 0.054 | 35% | 48% |
| 1341 | 1415 | 20442 | 20218 | -3 | 0.0 | 95% | 96% |
| 1425 | 2533 | 18566 | 15240 | -1 | 0.0 | 98% | 99% |
| 2534 | 3349 | 15178 | 12704 | -2 | 0.0 | 92% | 95% |
Summary of Gene C: Gene C appears to be the D. ananassae ortholog of the D. melanogaster gene Ten-m. The gene is on the minus strand (all matching reading frames are -1, -2, or -3). The 5' end of the gene does not appear to be on the fosmid. A view of D. melanogaster Ten-m from FlyBase GBrowse is shown below.
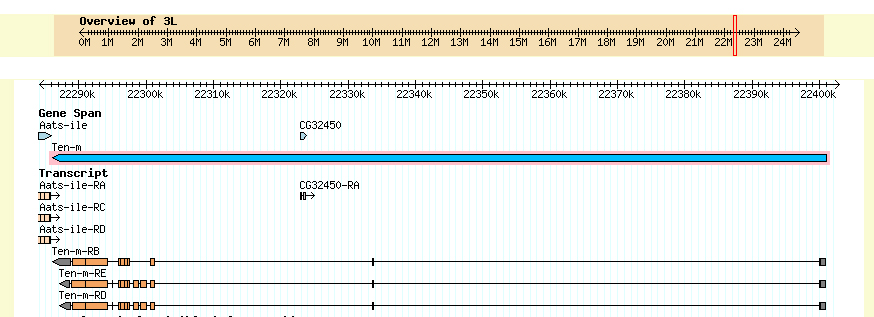
The D. melanogaster Ten-m gene has three isoforms derived from alternative splicing. The first and second introns are very large (approximately 70 kb and 30 kb, respectively). If the Ten-m gene of D. ananassae has a similar structure, the 5' end cannot be on the fosmid, which is only 44 kb.
To confirm this, I used the Gene Record Finder at GEP. This returns the following protein sequences for the first three exons of D. melanogaster Ten-m:
>Ten-m:13_536_0 MNPYEYESTLDCRDVGGGPTPAHAHPHAQGRTLPMSGHGRPTTDLGPVHG SQTLQHQNQQNLQAAQAAAQSSHYDYEYQHLAHRPPDTANNTAQRTHGRQ >Ten-m:12_536_2 FLLEGVTPTAPPDVPPRNPTMSRMQNGRLTVNNPNDADFEPSCLVRTPSG NVYIPSGNL >Ten-m:11_536_2 INKGSPIDFKSGSACSTPTKDTLKGYERSTQGCMGPVLPQRSVMNGLPAH HYSAPMNFRKDLVARCSSPWFGIGSISVLFAFVVMLILLTTTGVIKWNQS PPCSVLVGNEASEVTAAKSTNTDLSKLHNSSVRAKNGQGIGLAQGQSGLG AAGVGSGGGSSAATVTTATSNSGTAQGLQSTSASAEATSSAATSSSQSSL TPSLSSSLANANNG
These three sequences were used as subject sequences in a bl2seq search with BLASTX using fosmid_1475K17.fasta as the query sequence.
The first exon gives no significant match (E = 0.49). The second exon gives no significant match (E = 0.045). The third exon gives a significant match (E = 7e-66) to 25765 - 25316 on the fosmid, corresponding to the first segment of the fosmid that aligns with NP_001262211.1 in the analysis above. This shows that the third exon of Ten-m is the most 5' exon encoded on the fosmid.
Gene A. The eighth hit on the description list is HEM-protein. The coordinates of the alignment are shown in the table below.
| NP_524214.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 394 | 1121 | 3703 | 1520 | -2 | 0.0 | 99% | 98% |
| 1 | 395 | 4943 | 3759 | -1 | 0.0 | 97% | 98% |
Reordering these in order of the segments of NP_524214.1 gives the results shown below.
| NP_524214.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 1 | 395 | 4943 | 3759 | -1 | 0.0 | 97% | 98% |
| 394 | 1121 | 3703 | 1520 | -2 | 0.0 | 99% | 98% |
Summary of Gene A: Gene A is the ortholog of the D. melanogaster Hem gene. The D. ananassae Hem gene is on the minus strand (the aligning segments are in frames -1 and -2). The gene has two exons. NP_524214.1 has 1126 amino acids, so the entire Hem gene is on the fosmid.
Gene B. The ninth hit on the description list is Isoleucyl-tRNA synthetase. The coordinates of the alignment are shown in the table below.
| NP_730716.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 50 | 467 | 5833 | 7086 | +1 | 0.0 | 96% | 98% |
| 462 | 749 | 7121 | 7984 | +2 | 0.0 | 96% | 97% |
| 914 | 1228 | 8600 | 9541 | +2 | 0.0 | 77% | 87% |
| 750 | 918 | 8047 | 8553 | +1 | 0.0 | 91% | 95% |
| 1 | 58 | 5621 | 5806 | +2 | 2e-19 | 76% | 82% |
Reordering these in order of the segments of NP_730716.1 gives the results shown below.
| NP_730716.1 | fosmid | alignment | |||||
| start | end | start | end | frame | E | identity | positive |
| 1 | 58 | 5621 | 5806 | +2 | 2e-19 | 76% | 82% |
| 50 | 467 | 5833 | 7086 | +1 | 0.0 | 96% | 98% |
| 462 | 749 | 7121 | 7984 | +2 | 0.0 | 96% | 97% |
| 750 | 918 | 8047 | 8553 | +1 | 0.0 | 91% | 95% |
| 914 | 1228 | 8600 | 9541 | +2 | 0.0 | 77% | 87% |
Summary of Gene B: Gene B is the ortholog of D. melanogaster Aats-ile gene. The gene is on the + strand (all matching reading frames are +1 or +2). The gene has five exons. NP_730716.1 has 1229 amino acids, so the entire Aats-ile gene appears to be on the fosmid.
The project folder from GEP contains the following analysis (analysis/BLASTresults/dmel_translation_548_fosmid_1475K17.blx):
Smallest
Sum
High Probability
Sequences producing High-scoring Segment Pairs: Score P(N) N
Ten-m-PE FlyBase:FBpp0303192 5823 0. 11
Ten-m-PD FlyBase:FBpp0297244 5823 0. 11
Ten-m-PB FlyBase:FBpp0078161 5823 0. 8
Hem-PA FlyBase:FBpp0078162 3929 0. 2
Aats-ile-PA FlyBase:FBpp0078150 2289 0. 5
Aats-ile-PC FlyBase:FBpp0078151 2289 0. 5
Aats-ile-PD FlyBase:FBpp0078152 2289 0. 5
Ten-a-PE FlyBase:FBpp0289137 1374 0. 16
Ten-a-PH FlyBase:FBpp0289439 1374 0. 15
Ten-a-PI FlyBase:FBpp0289440 1374 0. 15
Ten-a-PJ FlyBase:FBpp0289441 1374 0. 15
Ten-a-PN FlyBase:FBpp0301779 1374 0. 15
Ten-a-PD FlyBase:FBpp0289136 1374 0. 15
Ten-a-PF FlyBase:FBpp0289138 1374 0. 15
Ten-a-PK FlyBase:FBpp0300541 1374 0. 15
Ten-a-PL FlyBase:FBpp0300542 1374 0. 15
Ten-a-PM FlyBase:FBpp0300543 1374 0. 15
Ten-a-PG FlyBase:FBpp0289438 1374 0. 14
CG5414-PB FlyBase:FBpp0291534 229 5.2e-42 6
Sgs1-PA FlyBase:FBpp0077084 81 9.4e-18 14
drpr-PE FlyBase:FBpp0306204 73 3.4e-14 11
drpr-PF FlyBase:FBpp0306205 77 9.9e-14 9
drpr-PB FlyBase:FBpp0072681 77 1.6e-13 9
crb-PC FlyBase:FBpp0293268 101 1.7e-13 6
crb-PB FlyBase:FBpp0110307 101 2.6e-13 6
crb-PD FlyBase:FBpp0306945 101 2.7e-13 6
CG5660-PA FlyBase:FBpp0076287 113 9.1e-13 6
C901-PA FlyBase:FBpp0073256 118 1.5e-12 3
drpr-PA FlyBase:FBpp0072680 110 8.3e-12 7
drpr-PC FlyBase:FBpp0301579 110 1.6e-11 6
crb-PA FlyBase:FBpp0083987 101 2.7e-11 8
Muc96D-PA FlyBase:FBpp0084219 80 5.4e-11 10
I combined isoforms into single genes, then identified the segment of the fosmid aligning to each sequence, as summarized in the table below. A representative protein sequence (NP_xxxxx) from D. melanogaster was chosen for each gene (only one isoform for genes with multiple isoforms) and used as the subject sequence for a bl2seq search with BLASTX using fosmid_1475K17.fasta as the query sequence. All aligning segments were combined to give a coordinate range on the fosmid. The E value for the highest scoring segment is shown.
Two of the proteins in the report do not significantly align with the translation of the fosmid in this analysis. All of the remaining alignments correspond to the coordinate range for two of the genes identified above (Aats-ile and Ten-m), aligning with lower scores, indicating homology to these genes. These are not new genes missed by the BLASTX analysis above.
| Dmel gene | Representative protein sequence | fosmid | E | Conclusion | ||
| start | end | strand | ||||
| Ten-m | NP_001262211.1 | 25759 | 12704 | minus | 0.0 | Dana Ten-m (see above) |
| Hem | NP_524214.1 | 4943 | 1520 | minus | 0.0 | Dana Hem (see above) |
| Aats-ile | NP_730716.1 | 5621 | 9541 | plus | 0.0 | Dana Aats-ile (see above) |
| Ten-a | NP_001138189.1 | 21552 | 12938 | minus | 0.0 | Homology to Ten-m |
| CG5414 | NP_648837.2 | 5836 | 7966 | plus | 2e-48 | Homology to Aats-ile |
| Sgs-1 | NP_523475.3 | 14537 | 14632 | plus | 2.2 | Not significant |
| drpr | NP_001261276.1 | 21405 | 20639 | minus | 9e-13 | Homology to Ten-m |
| crb | NP_001247284.1 | 21429 | 20528 | minus | 1e-06 | Homology to Ten-m |
| CG5660 | NP_648268.1 | 5848 | 7657 | plus | 2e-18 | Homology to Aats-ile |
| C901 | NP_572673.1 | 21399 | 20525 | minus | 2e-13 | Homology to Ten-m |
| Muc96D | NP_733106.2 | 40701 | 40654 | minus | 2.8 | Not significant |
FlyBase BLAST makes the analysis of the fosmid easier in some ways. I used the link to FlyBase BLAST from the Tools page of the course website.
I set the Database to Annotated proteins (AA), the Program to BLASTX, and uploaded the fosmid sequence. I restricted the species to D. melanogaster and clicked BLAST.
The graphic output is shown below. Notice that each hit is labeled, unlike the results at NCBI BLAST.
The summary table is also useful, shown below.
The GENSCAN results from analysis/Genefinder/Genscan in the project folder predict five proteins on the fosmid:
>fosmid_1475K17|GENSCAN_predicted_peptide_1|1126_aa MARPIFPNQQKIAEKLIILNDRGLGILTRIYNIKKACGDTKSKPGFLSEKSLESSIKFIV KRFPNIDVKGLNAIVNIKAEIIKSLSLYYHTFVDLLDFKDNVCELLTTMDACQIHLDITL NFELTKNYLDLVVTYVSLMIVLSRVEDRKAVLGLYNAAYELQNNQADTGFPRLGQMILDY EVPLKKLAEEFIPHQRLLANALRSLTSIYALRNLPADKWREMQKLSLVGNPAILLKAVRT ETMSCEYISLEAMDRWIIFGLLLNHQMLGQYPEVNKIWISALESSWVVALFRDEVLQIHQ YIQSTFDGIKGYSKRISEVKDAYNTAVQKAAHMHRERRKFLRTALKELALIMTDQPGLLG PKAIFIFIGLCLARDEILWLLRHNDNPPPVKNKGKSNEDLVDRQLPELLFHMEELRALVR KYSQVMQRYYVQYLSGFDATDLNIRMQSLQMCPEDESIIFSSLYNIAASLTVKQVEDNEL FYFRPFRLDWFRLQTYMSVGKAALRITEHIELARLLDSMVFHTRVVDNLDEILVETSDLS IFCFYNKMFDDQFHMCLEFPAQNRYIIAFPLICSHFQNCTHEMCPEERHHIRERSLSVVN IFLEEMAKEAKNIITTICDEQCTMADALLPKHCAKILSVQSARKKKDKAKSKHFDDIRKP GDESYRKTREDLTTMDKLHMALTELCFAINYCPTVNVWEFAFAPREYLCQNLEHRFSRDL VGMVMFNQETMEIAKPSELLASVRAYMNVLQTVENYVHIDITRVFNNCLLQQTQALDSHG EKTIAALYNTWYSEVLLRRVSAGNIVFSINQKAFVPISPEGWVPFNPQEFSDLNELRALA ELVGPYGIKTLNETLMWHIANQVQELKSLVVTNKEVLITLRTSFDKPEVMKEQFKRLQDV DRVLQRMTIIGVIICFRNLVHEALVDVLDKRIPFLLSSVKDFQEHLPGGDQIRVASEMAS AAGLLCKVDPTLATTLKSKKPEFDEGEHLTACLLMVFVAVSIPKLARNENSFYRATIDGH SNNTHCMAAAINNIFGALFTICGQNDMEDRMKEFLALASSSLLRLGQESDKEATRNRESI YLLLDEIVKQSPFLTMDLLESCFPYVLIRNAYHGVYKQEQILGLVL >fosmid_1475K17|GENSCAN_predicted_peptide_2|1228_aa MGKKLERSDVCRVPENINFPAEEENVLQRWREENVFERCSQLSKGKPKYTFYDGPPFATG LPHYGHILAGTIKDIVTRYAYQQGYHVDRRFGWDCHGLPVEFEIDKLMNIRGPEDVAKMG ITAYNAECRKIVMRYADEWENIVTRVGRWIDFKNDYKTLYPWYMESIWWIFKQLYDKGLV YQGVKVMPYSTACTTSLSNFEANQNYKEVVDPCVVIALEAVSLPNTFFLVWTTTPWTLPS NFACCVHPTMTYVKVRDVKSDRLFILAESRLSYVYKTEAEYEVKDKFAGKTLKDLHYKPL FPYFAKRGAEVKAYRVLVDEYVTEDSGTGIVHNAPYFGEDDYRVCLAAGLITKSSEVLCP VDEAGRFTKEASDFEGQYVKDADKQIMAVLKTRGNLVSSGQVKHSYPFCWRSDTPLIYKA VPSWFVRVEHMSKNLLTCSSQTYWVPDFVKEKRFGNWLREARDWAISRNRYWGTPIPIWR SPNGDETIVIGSIKQLAELSGVQVEDLHRESIDHIEIPSAVPGNPPLRRIAPVFDCWFES GSMPFAQQHFPFENEKDFMNNFPADFIAEGIDQTRGWFYTLLVISTALFNKAPFKNLIAS GLVLAADGQKMSKRKKNYPDPMEVVHKYGADALRLYLINSPVVRAESLRFKEEGVRDIIK DVFLPWYNAYRFLLQNIARYEKEDLGGKGQYIYERERHLKNMDKASVIDVWILSFKESLL QFFAEEMKMYRLYTVVPRLTKFIDQLTNWYVRLNRRRIKGELGAEQCIQSLDTLYDVLYT MVKMMAPFTPYLTEYIFQRLVLFQPPGSLEHADSVHYQMMPVSQSKFIRNDIERSVSLMQ SVVELGRVMRDRRTLPVKYPVSEIIVIHKDAKVLEAVKNLQDFILSELNVRKLTLSSDKE KYGVTLRAEPDHKTLGQRLKGNFKAVMAAIKALKDDEIQKQVAQGYFNILDQRIELDEVR VIYCTSEQVGGHFEAHSDNEVLVLLDMTPNEELLEEGLAREVINRVQKLKKKAQLIPTDP VLIFHELEANSTKKETLETQAQLKKVLSSYSDMIKTAIKSDFGPFSAEKSAKKRVIASEL VDLKGIPLKLTICSTEDLQLPNLPFLNVALAEDLKPRFGNGDKASLFLQHNASKQIISLP QLRSEIEILFGLYGVNFNIYVVDQKSNAKELTSIDKSLNGKLLVLSRGPEALKSKASFDV PSSPYSKFVNKGSGTAAFIENPKGTTLN >fosmid_1475K17|GENSCAN_predicted_peptide_3|3291_aa MRHVAVQTNECARKYAIEFKYCKCMSRTLGPGRGEQQQRNNWRYMFFAAVPQPPIPANDT RFQTTLRRCKKGGYGMRIRMRTRSPIIYPNTHLIRTRGGAQVACYINKGSPIDFKSGSAC STPTKETLKGGYDRGTQGCMGPVLPPRSVMNGLPSHHYSAPMNFRKDLAARCSSPWVGVA AISALVLVVLMLILLTTFGGLHWTQSAPCTVLVGNEASEVTAAKSTNTDLSKLHNSATRS KNGQAIGPVPGQYGSGGGMGSSTATVTAATSNSGTAQGLQSTSASAEASSSAATTSSQSS LTPSTSLSSSLANANNGGRDILTRMAADGAGKNKRRNRRSMDVAENGGDVATDETFSNFI TIESLNREQTGEYFATTPARKLQEVERSSSDRTSFGINGVLSPQGDEEVEDITSDYVYED EPVPDTSPATQRPRTRQQFGKSLNSNLRSAAKTLVNKKTKYEGEAGKNIRLEQEQKLEAF IEAGMTLESTTTTATRATTTTESGTSTFVAVIDDDNQSDSSSSGAVPPVLTVLRSDTDDI VEINTALPEPTEGSILAPFPRSSMANDFQIKGKAVSESGQEKPATDDNNNERDLADNYEL KPEEPAASATPLQGINQVHSTFLAGEINKSESDFMVNDMASQFEDIDIVKLGEAPSSHEE AVYKTSSSKDKAVPMAPAQPEAIENEVLKDQDEARQVPLHLRPLKPYVSETIDQPGRRIL VNLTIATDDGPDNVYTLHVEVPTGGGPHTIKEVLTHEKPPQQADHQAENCVPEPPPRMPD CPCSCLPPPAPIYLDDRGDSGDSGSAPPVDTDSAPLASTTNGASTSPPLETATILGDRHS EKDDHGVGNGNSSTVEGESTASSCATNTPSTEIDNHIDAFHTEPPVGGGEPFACPDVMPV LILEGARTFPARSFPPDGTTFGQISLGQKLTKEIQPYSYWNMQFYQSEPAYVKFDYTIPR GASIGVYGRRNALPTHTQYHFKEVLSGFSASTRTARAAHLSITREVTRYMEPGHWFMSLY NDDGDVQELTFYAAVAEDMTQNCPNGCSGNGQCLLGHCQCNPGFGGHDCSESVCPVLCSQ HGEYTNGECICNPGWKGKECSLRHDECEVADCNGHGHCVSGKCQCMRGYKGKFCEEVDCP HPNCSGHGFCADGTCICKKGWKGPDCATMDQDALQCLPDCSGHGTFDLDTQTCTCEAKWS GDDCSKELCDLDCGQHGRCEGDACACDPEWGGEYCNTRLCDTRCNEHGQCKNGTCLCVTG WNGKHCTIEGCPNSCAGHGQCRVSGEGQWECRCYEGWDGPDCGIALELNCGDSKDNDKDG LVDCEDPECCASHVCKTSQLCVSAPKPIDVLLRKQPPAITASFFERMKFLIDESSLQNYA KLETFNESRSAVIRGRVVTSLGMGLVGVRVSTTTLLEGFTLTRDDGWFDLMVNGGGAVTL QFGRAPFRPQSRIVQVPWNEVVIIDGVVMSMSEEKGLATTTTHTCFAHDYDLMKPVVLAS WKHGFQGACPDRSAILAESQVIQESLQIPGTGLNLVYHSSRAAGYLSTIKLQLTPDNIPP TLHLIHLRITIEGILFERVFEADPGIKFTYAWNRLNIYRQRVYGVTTAVVKVGYQYTDCT DIVWDIQTTKLSGHDMSISEVGGWNLDIHHRYNFHEGILQKGDGSNIYLRNKPRIILTTM GDGHQRPLECPDCDGLATKQRLLAPVALAAAPDGSLFVGDFNYIRRIMSDGSIRTVVKLN ATRVSYRYHMALSPLDGTLYVSDPESHQIIRVRDTNNYSQPELNWEAVVGSGERCLPGDE AHCGDGALAKDAKLAYPKGIAISSDNILYFADGTNIRMVDRDGIVSTLIGNHMHKSHWKP IPCEGTLKLEEMHLRWPTELAVSPMDNTLHIIDDHMILRMTPDGRVRVISGRPLHCATAS TAYDTDLATHATLVMPQSIAFGPLGELYVAESDSQRINRVRVIGTDGRIAPFAGAESKCN CLERGCDCFEAEHYLATSAKFNTIAALSVTPDGHVHIADQANYRIRSVMSSIPEASPSRE YEIYAPDMQEIYIFNRFGQHVSTRNILTGETTYVFTYNVNTSNGKLSTVTDAAGNKVFLL RDYTSQVNSIENTKGQKCRLRMTRMKMLHELSTPDNYNVTYEYHGPTGLLKTKLDSTGRS YVYNYDEFGRLTSAVTPTGRVIELSFDLSVKGAQVKVSENAQKEQSLLIQGATVTVRNGA AESRTSVDMDGSTTSITPWGHNVQMEVAPYTILAEQSPLLGESYPVPAKQRTEIAGDLAN RFEWRYFVRRQQPLQAGKQSKGAPRPVTEVGRKLRVNGDNVLTLEYDRETQSVVVLVDDK QELLNVTYDRTSRPISFRPQSGDYADVDLEYDRFGRLVSWKWGVLQEAYSFDRNGRLNEI KYGDGSTMVYAFKDMFGSLPLKVTTPRRSDYLLQYDDAGALQSLTTPRGHIHAFSLQTSL GFFKYQYFSPINRHPFEILYNDEGQILAKIHPHQSGKVAFVYDAAGRLETILAGLSSTHY TYQDTTSLVKTVEVQEPGFELRREFKYHAGILKDEKLRFGSKNSLASAHYKYAYDGNARL SGIEMAIDDKELPTTRYKYSQNLGQLEVVQDLKITRNAFNRTVIQDSAKQFFAIVDYDQH GRVKSVLMNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNADGHVVEVLGTNN WKYLYDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGFVVRRGEQKYRYNN RGQLIHAFERERFQSWYYYDDRSRLVAWHDNQGNTTQYYYANPRTPHLVTHAHFPKLART MKFFYDDRDMLIAMENADQRYYVATDQNGSPLAFFDLNGGIAKELKRTPFGRIIKDTKPD FFVPIDFHGGLIDPHTKLIYTEQRQYDPHVGQWMTPQWETLATEMSHPTDVFIYRYHNND PINPNRPQNYMIDLDAWLQLFGYDLDNMQSRRYTKLAQYTPQASIKSNMLAPDFGVISGL ECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRISYRRGVFGEGVLLSRIGGRALVSVV DGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVKDNVLKLRDDNEELRRLGGMFNIS THEVSDHGGSAAKELRLHGPDAVVIVKYGVDPEQERHRILKHAHKRAVERAWELEKQLVA AGFQGRGDWTEEEKEELVQHGDVDGWIGIDIHSIHKYPQLADDPGNVAFQRDAKRKRRKT GNSHRSASSRRQMKFGELSALYDYDCNEQLVFNVENSLENQISRRRRNEKY >fosmid_1475K17|GENSCAN_predicted_peptide_4|111_aa MEKKPSNNGRFNGRLSEASAIINGRNNSLLRRRLLATTEGELVGGERGGREVAPWTHIID QNFTDYKAINGIDYSISKTDRNFAFAAQFLRVQPPSRKFNIIALGLAVIAH >fosmid_1475K17|GENSCAN_predicted_peptide_5|46_aa XYGTCLCEHCQLDELLLRQLIALPPDNWWDSQRFYVLIVIIVTAAL
Each protein was used as a query sequence of a BLASTP search of Non-redundant protein sequences (nr) restricted to Drosophila melanogaster. All BLAST parameters were left at the default settings. The results are summarized in the table below. Peptides 4 and 5 do not produce significant alignments with D. melanogaster proteins.
| Query | Top hit | E | Coverage | Max identity | |
| Accession | Gene | ||||
| GENSCAN_predicted_peptide_1 | NP_524214.1 | Hem | 0.0 | 100% | 98% |
| GENSCAN_predicted_peptide_2 | NP_730716.1 | Aats-ile | 0.0 | 99% | 91% |
| GENSCAN_predicted_peptide_3 | NP_001262211.1 | Ten-m | 0.0 | 97% | 86% |
| GENSCAN_predicted_peptide_4 | No significant similarity found. | ||||
| GENSCAN_predicted_peptide_5 | NP_609068.2 | Ttll3B | 3.2 | 60% | 39% |
Because no significant matches were found with peptides 4 and 5, I repeated the BLASTP search of nr with the species restriction turned off.
The top hit for peptide 4 was WP_004022292.1 with an E value of 2.0 (not significnat).
The top hit for peptide 5 was WP_005224522.1 (conjugal transfer protein TraB [Marichromatium purpuratum]) with an E value of 7e-06 (marginally significant). The alignment is shown below.
Query 3 GTCLCEHCQLDELLLRQLIALPPDNWWDSQRFYVLIVIIVTA 44
G CL E + E ++ +L LPP N W + +VL+V+I+
Sbjct 229 GRCLAEEDESPEPVIAELERLPPPNPWPRRLPWVLVVLILAG 270
Summary of GENSCAN analysis: GENSCAN identified three genes found by the BLASTX search (Hem, Aats-ile, and Ten-m). It made two additional predictions that are invalid.
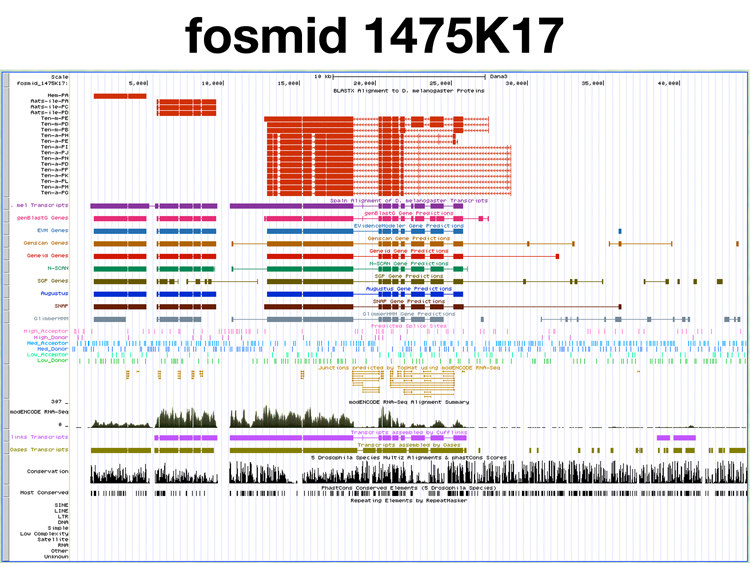
Here is a view of fosmid 1475K17 in the UCSC Genome Browser at GEP.
BLASTX Alignment of D. melanogaster proteins. The BLASTX track at the top of the image shows alignments to three distinct regions, as was seen in the prior BLASTX analaysis. The leftmost D. ananassae gene, Hem, aligns to the protein product of the D. melanogaster Hem gene and no other sequences. The middle D. ananassae gene, Aats-ile, aligns to three D. melanogaster isoforms produced by the D. melanogaster Aats-ile gene. The rightmost D. ananassae gene, Ten-m, aligns to isoforms produced by the D. melanogaster Ten-m gene. In addition, the protein isoforms of the closely-related but distinct D. melanogaster Ten-a gene align to the D. ananassae Ten-m gene. These are the same results seen when the fosmid is used as a query sequence in a BLASTX search of D. melanogaster proteins. There are three genes on the fosmid: the D. ananassae orthologs of Hem, Aats-ile, and Ten-m.
GENSCAN predictions. Starting at the left of the fosmid, the first three GENSCAN predictions align to the D. ananassae orthologs of Hem, Aats-ile, and Ten-m, as shown in the previous analysis. The third GENSCAN prediction contains one additional 3' exon and two additional 5' exons in the Ten-m gene that are not supported by BLASTX analysis. The fourth and fifth GENSCAN predicitons do not align to sequences predicted by BLASTX analysis to encode proteins. The first three GENSCAN predictions are congruent with predictions from other gene-finding programs, while the invalid fourth and fifth predictions do not match predictions from other gene-finding programs, with the exception of one of the exons of GENSCAN peptide 5, also predicted by SNAP.
modENCODE RNA-Seq. Transcripts aligning to Hem, Aats-ile, and Ten-m are seen. There is little evidence of RNA sequences present in mRNA elsewhere on the fosmid.
Conservation. The exons of Hem, Aats-ile, and Ten-m are clearly conserved. The intergenic regions between Hem and Aats-ile, and between Aats-ile and Ten-m are not conserved. There is considerable sequence conservation upstream of the rightmost (5') exon in Ten-m; this region is known to be a Ten-m intron, separating the third exon of Ten-m from the second exon, which is not on the fosmid.