Summary
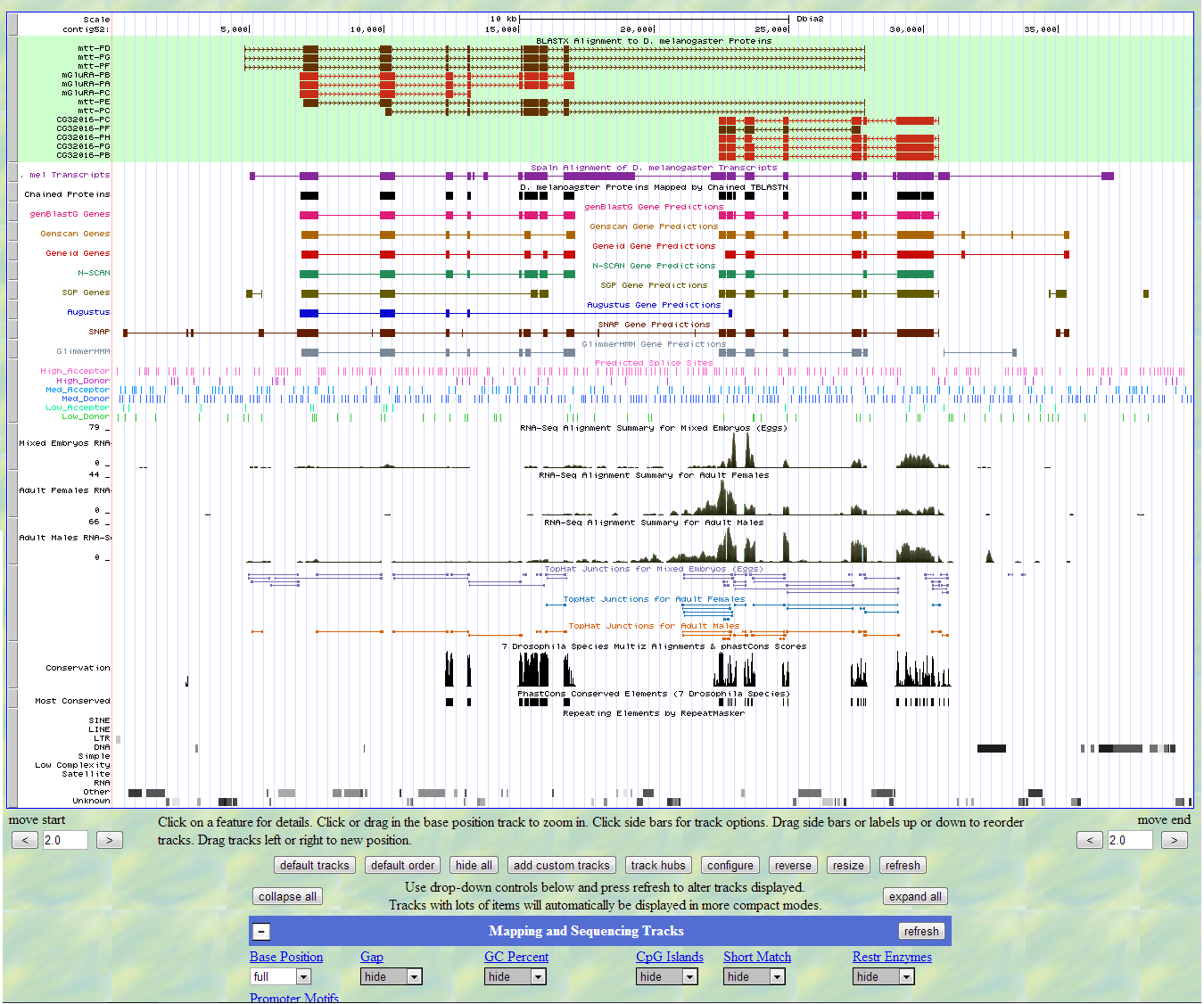
D. biarmipes contig 52 contains two genes. The orthologs of the D. melanogaster genes CG32016 and mGLuRA. GENSCAN predicts two peptides on the contig. Blastx predicts two genes as well.
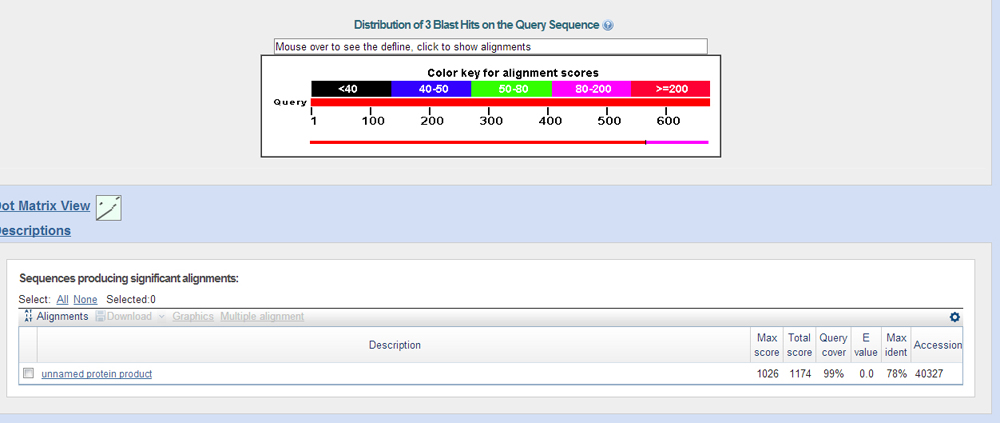
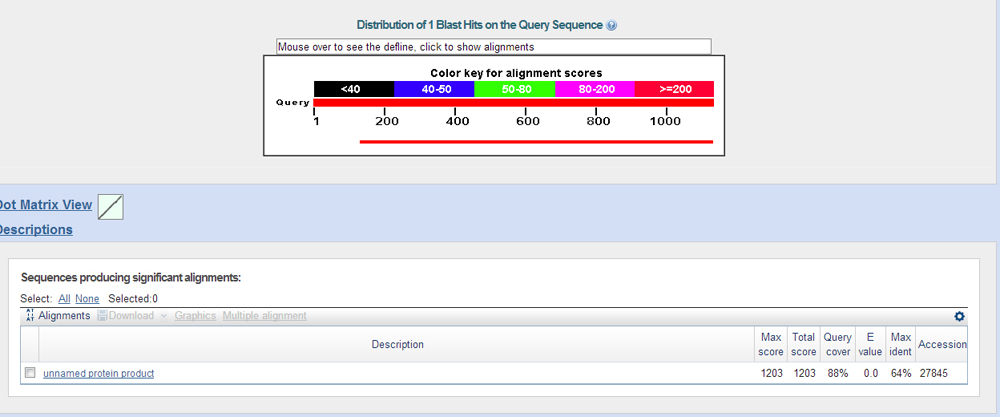
Blastx

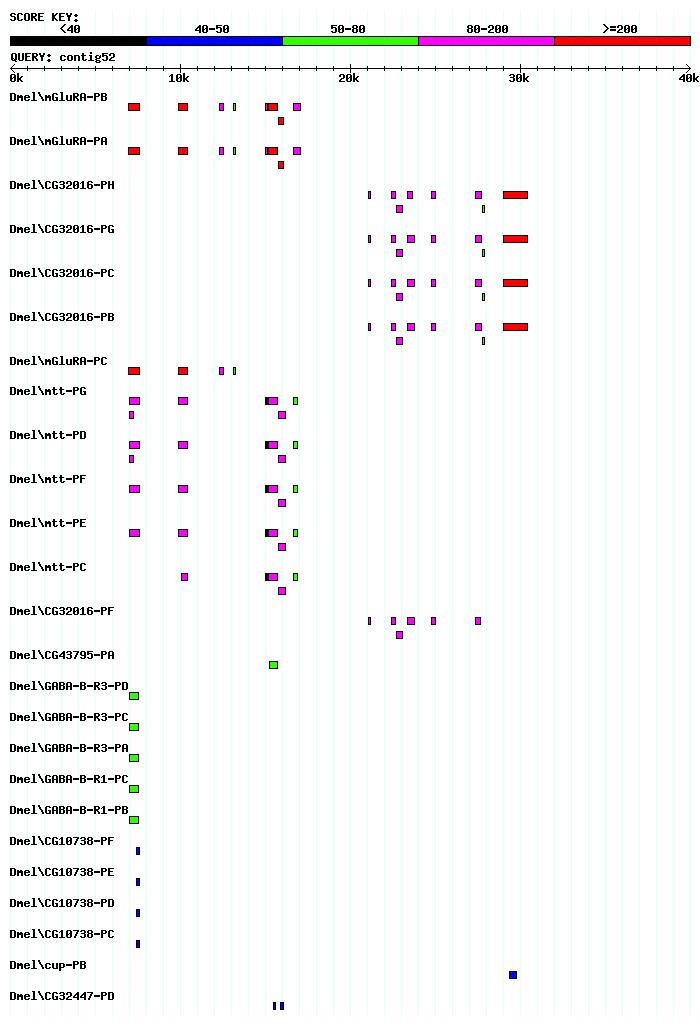
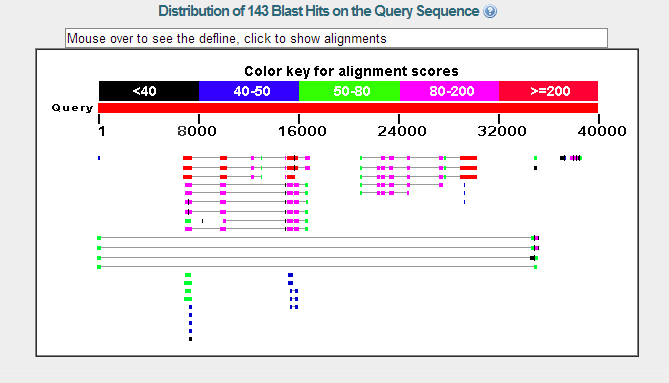
The blastx shows two genes. The gene on the right is the mGLuRA gene. The gene on the left is the CG32016 gene. The two different blasts that were run both agree on the two genes that are present on this contig.

The E-Values for the blasts also agree that these are the two genes on the contig 52.
Alignments
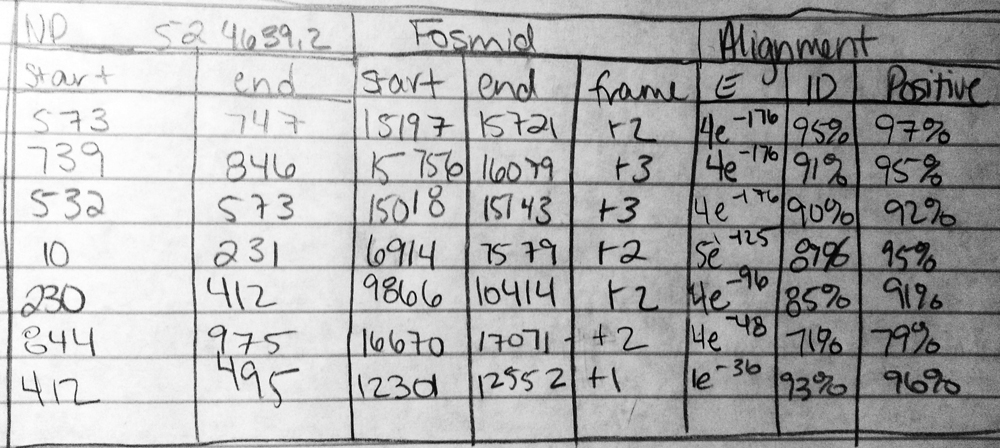
The first 7 alignments of mGLuRA (NP_524639.2) are shown above.

The first four alignments of CG32016 Isoform B (NP_001259078.1) are shown above.
GenScan
GenScan predicted two genes on the contig.
Gene 1: MLMLSPVANLKSLNNVHTQDTVSVSLPGDIILGGLFPVHEKGEGAPCGPKVYNRGVQRLE AMLYAIDRVNNDSNILPGITIGVHILDTCSRDTYALNQSLQFVRASLNNLDTSVFECSDS SSPQIRKNASSGPVFGVIGGSYSSVSLQVANLLRLFHIPQISPASTAKTLSDKSRFDLFA RTVPPDTFQSVALVDILKNFNWSYVSTIHSEGSYGEYGIEAFHKEATERHVCIAVAEKVP SAADDKVFDLIIGKLQKKPNARGVVLFTRAEDARRILQAAKRANLSQPFHWIASDGWGKQ QKLLEGLEDIAEGAITVELQSEIIEDFDHYMMQLTPETNQRNPWFAEYWEDTFNCILEPV SDQTNSPTSIDSTEIKIATKSKTTCEDSFRLSEKVGYEQESKTQFVVDAVYAFAYALHNL HNDRCNTQSDQTSEQRKHHHNLAGSEVKFDRQGDGLARYDILNYQRLENSSGYQYKGDTC CWICDSCESFEYVYDEFTCKDCGPGLWPYADKLSCFALDIQYMRWNSLFALIPMAIAIFG IAVTIIVMLLFAKNHDTPLVYILVFHPDKNVRKLTMNSTVYRRSAATGAQGAPSSSVYSR TQAGNTVPTGGALGTTASSALQTQNSSNLDEPSGQSAVVHKSSDYSNGEFMPEECECAEA FCNRVKN
Gene 2: MYRVSGSPTYLPLNGCSPKRQDRQFGYVALEDSLSGFYGDHSDFVVLSGVHMPTFPKRFM GTFRRDGSVSDTAIEGKVTAAKFRRCKRIEASASSSSALFVKYLVIVESISSLMESTAEI VLQGAVLKIYLGTPYTRADLLALRYEGKSRQRPHCRNRTELHTLGFWKVNLNAVSLSVAN NYSNQNKNRLSPEADSSSLNCSNSASMSSRRAMRNRERANNYYQRFVPTESLQMCGEDKD KDKDATTQGQSFKSPVIDHRSISSSHLMPAFAKRRIAASSGTNNGETNETSVSTCDAAPS AHQRRESKGKVPSSPNRKSSELDMAETRLNYVHQEHDQYMSSSPTFSTSRQERRIGSGRL LPRSDNWEFKSQKTKEPNTETEKDASPNGSGGASVANQQNQNQHHQRVFSGRLVDRVTEH TDRRFQNDTKRSVDRQGVGNRRISNKEPCSNQNRGKRANSYHVHEEPEWFSAGPTSQLET IDLHGFDDLDNNEDRSEMDNEKFLQIDTNLAAQTTIDEASRRNSNVSLNLSDAYQSDDII DTGENILKCIQNSSELCKQNQNEQSQFQCSQSTESEFNFDAFLNMHPLDNSLMNNDENEK GEATGTSRFSRWFRHKETANNNELPGLQDFNAQEKIGIPSVKDLEAQMTKVDMRPDYVNT VGGPFSQVVQAEKPIPRDTEGFKKLLQQLGYQSRQHHPGNDVYHIINHSNITNHDHLESN QQHKINNCHSQHSALSVHGPNIPSNSHIFAQKRLETHHLMQSLICGDVSLDFLEKELGNP STAPSTKEVIASVLREYSHNKRNPVSIGDHKMFTHSTFLQAQPVHPHYSEELISQNTANH AMNPLITHGNSPTPLAFTPTSVLRKMTADKETTQTPSSSHSQQPQYQMHPQHAKQLSETQ PTAPIAVQPRMILGGGNFVIGPNNQPISPNLQQCRNQQGIKWTPGRPILKGGLNSMPQPN PALTFTTHKIEMQPVHSHHQQLQQQQTQHRFKSGQTVESILNTEHVHQNIPSPVGWHQLF LQHQQHQSRQQPRHRMLYGEMHRQSNPQMSSPVPGIPDSSDSGNVIKNNSNASPGYPRDE RMPSPTNNQLAQWFSPELLAKASAGKLPLLNMNQALSLEEFERSIQHSSAVVHN
After doing a protein blast of the GenScan predictions against the fasta's of the proteins from the blast x results, you see that these are indeed the genes on the contig.
Genome Browser at UCSC at GEP
After running the contig through the Genome Browser, it is clear that it also predicts at least two genes. It also shows that there are a lot of repeats in the contig as well as some conservation. Most of the different sites agree on the same genes present.